1E3F
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-06-14 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8QIF
 
 | | CrPhotLOV1 light state structure 12.5 ms (10-15 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
6H5Z
 
 | | Ferric murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
7YWK
 
 | | Crystal structure of an engineered TycA variant, TycApPLA, in complex with AMP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Mittl, P, Camus, A, Truong, G, Markert, G, Hilvert, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Reprogramming Nonribosomal Peptide Synthetases for Site-Specific Insertion of alpha-Hydroxy Acids.
J.Am.Chem.Soc., 144, 2022
|
|
8QIV
 
 | | CrPhotLOV1 light state structure 87.5 ms (85-90 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 11, 2024
|
|
5M6V
 
 | |
6SNL
 
 | | (R)-selective amine transaminase from Exophiala sideris | | Descriptor: | CHLORIDE ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Telzerow, A, Hakansson, M, Steiner, K. | | Deposit date: | 2019-08-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | Expanding the Toolbox of R-Selective Amine Transaminases by Identification and Characterization of New Members.
Chembiochem, 22, 2021
|
|
5M79
 
 | |
1DNK
 
 | | THE X-RAY STRUCTURE OF THE DNASE I-D(GGTATACC)2 COMPLEX AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*GP*GP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3'), ... | | Authors: | Weston, S.A, Lahm, A, Suck, D. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution.
J.Mol.Biol., 226, 1992
|
|
7RSQ
 
 | | Cryo-EM structure of KIFBP core | | Descriptor: | KIF-binding protein | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
7A99
 
 | | Crystal structure of the Phe57Trp mutant of the arginine-bound form of domain 1 from TmArgBP | | Descriptor: | ACETATE ION, ARGININE, Amino acid ABC transporter, ... | | Authors: | Balasco, N, Vitagliano, L, Smaldone, G, Ruggiero, A. | | Deposit date: | 2020-09-01 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Development of a Protein Scaffold for Arginine Sensing Generated through the Dissection of the Arginine-Binding Protein from Thermotoga maritima .
Int J Mol Sci, 21, 2020
|
|
6O4I
 
 | | Structure of ALDH7A1 mutant E399D complexed with alpha-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
1DO5
 
 | | HUMAN COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE DOMAIN II | | Descriptor: | HUMAN COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE DOMAIN II, ZINC ION | | Authors: | Lamb, A.L, Wernimont, A.K, Pufahl, R.A, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 1999-12-18 | | Release date: | 2000-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the second domain of the human copper chaperone for superoxide dismutase.
Biochemistry, 39, 2000
|
|
7MXO
 
 | | CryoEM structure of human NKCC1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
8BDO
 
 | | VCB in complex with compound 21 | | Descriptor: | (2~{S},4~{R})-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-~{N}-[4-(4-methyl-1,3-thiazol-5-yl)phenoxy]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
5M8A
 
 | | Crystal structure of Eremococcus coleocola manganese transporter mutant E129A | | Descriptor: | Divalent metal cation transporter MntH | | Authors: | Manatschal, C, Ehrnstorfer, I.A, Arnold, F.M, Laederach, J, Dutzler, R. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family.
Nat Commun, 8, 2017
|
|
1E43
 
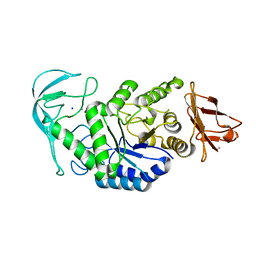 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.7A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
3OU3
 
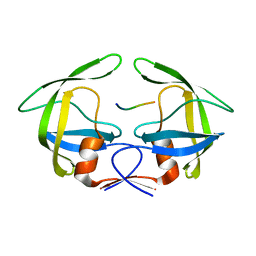 | | MDR769 HIV-1 protease complexed with PR/RT hepta-peptide | | Descriptor: | HIV-1 protease, PR/RT substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
4TNT
 
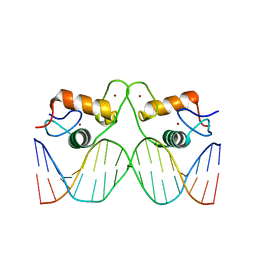 | | Structure of the human mineralocorticoid receptor in complex with DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), Mineralocorticoid receptor, ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2014-06-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3938 Å) | | Cite: | Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA.
Plos One, 9, 2014
|
|
3OUC
 
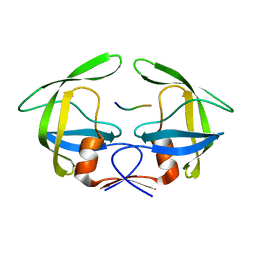 | | MDR769 HIV-1 protease complexed with p2/NC hepta-peptide | | Descriptor: | MDR HIV-1 protease, p2/NC substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
1DW9
 
 | | Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site | | Descriptor: | CHLORIDE ION, CYANATE LYASE, SULFATE ION | | Authors: | Walsh, M.A, Otwinowski, Z, Perrakis, A, Anderson, P.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 1999-12-03 | | Release date: | 2000-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Cyanase Reveals that a Novel Dimeric and Decameric Arrangement of Subunits is Required for Formation of the Enzyme Active Site
Structure, 8, 2000
|
|
7MH7
 
 | |
4WFT
 
 | | Crystal structure of tRNA-dihydrouridine(20) synthase dsRBD domain | | Descriptor: | tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
6OBN
 
 | | The crystal structure of coexpressed SDS22:PP1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8BDT
 
 | | Ternary complex between VCB, BRD4-BD2 and PROTAC 51 | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-3-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethylamino]-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-3-oxidanylidene-propyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A.A. | | Deposit date: | 2022-10-20 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
