8QDH
 
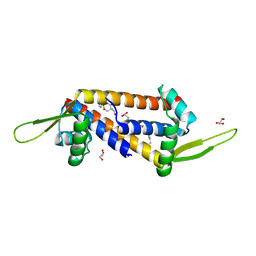 | | Engineered LmrR carrying a cyclic boronate ester formed between Tris and p-boronophenylalanine at position 89 | | Descriptor: | GLYCEROL, Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Longwitz, L, Leveson-Gower, R.B, Roelfes, G. | | Deposit date: | 2023-08-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Boron catalysis in a designer enzyme.
Nature, 629, 2024
|
|
3B3R
 
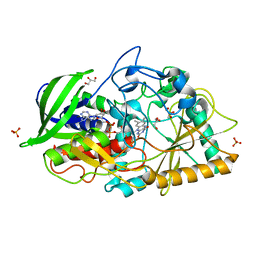 | | Crystal structure of Streptomyces cholesterol oxidase H447Q/E361Q mutant bound to glycerol (0.98A) | | Descriptor: | Cholesterol oxidase, FLAVIN-N7 PROTONATED-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lyubimov, A.Y, Heard, K, Tang, H, Sampson, N.S, Vrielink, A. | | Deposit date: | 2007-10-22 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Distortion of flavin geometry is linked to ligand binding in cholesterol oxidase
Protein Sci., 16, 2007
|
|
7M3Z
 
 | | Structure of TIM-3 in complex with N-(4-(8-chloro-2-mehtyl-5-oxo-5,6-dihydro-[1,2,4]triazolo[1,5-c]quinazolin-9-yl)-3-methylphenyl)methanesulfonamdide (compound 35) | | Descriptor: | CALCIUM ION, Hepatitis A virus cellular receptor 2, N-{4-[(4S,10aP)-8-chloro-2-methyl-5-oxo-5,6-dihydro[1,2,4]triazolo[1,5-c]quinazolin-9-yl]-3-methylphenyl}methanesulfonamide | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
7ME4
 
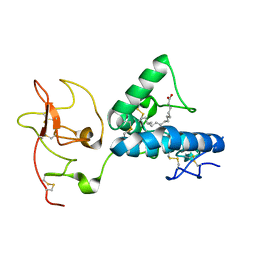 | | Structure of the extracellular WNT-binding module in Drosophila Ror2/Nrk | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOLEIC ACID, Tyrosine-protein kinase transmembrane receptor Ror2 | | Authors: | Mendrola, J.M, Shi, F, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
7QBJ
 
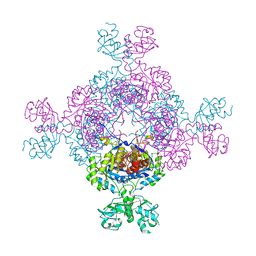 | | bacterial IMPDH chimera | | Descriptor: | Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
4M1Q
 
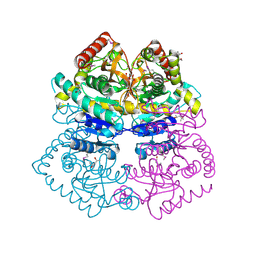 | | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-lactate dehydrogenase, PHOSPHATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-03 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus selenitireducens MLS10, NYSGRC Target 029814.
To be Published
|
|
3B6J
 
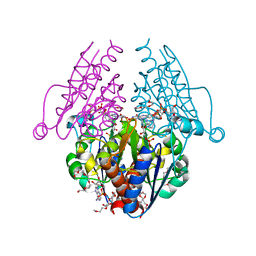 | | WrbA from Escherichia coli, NADH complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, Flavoprotein wrbA, ... | | Authors: | Andrade, S.L.A, Patridge, E.V, Ferry, J.G, Einsle, O. | | Deposit date: | 2007-10-29 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli.
J.Bacteriol., 189, 2007
|
|
7ME5
 
 | | Structure of the extracellular WNT-binding module in Drl-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tyrosine-protein kinase transmembrane receptor DRL-2 | | Authors: | Shi, F, Mendrola, J.M, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
8DRM
 
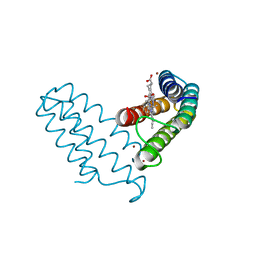 | |
5MUN
 
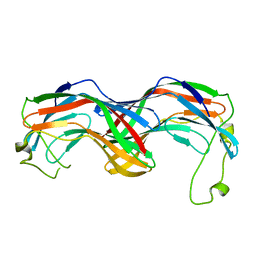 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
8DRJ
 
 | |
2PHW
 
 | | Pterocarpus angolensis lectin (PAL) in complex with Man-9 | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Garcia-Pino, A, Buts, L, Wyns, L, Imberty, A, Loris, R. | | Deposit date: | 2007-04-12 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How a plant lectin recognizes high mannose oligosaccharides
Plant Physiol., 144, 2007
|
|
6ZWO
 
 | | cryo-EM structure of human mTOR complex 2, focused on one half | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
2PIS
 
 | | Efforts toward Expansion of the Genetic Alphabet: Structure and Replication of Unnatural Base Pairs | | Descriptor: | DNA (5'-D(*CP*GP*(CBR)P*GP*AP*AP*(FFD)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Matsuda, S, Fillo, J.D, Henry, A.A, Wilkins, S.J, Rai, P, Dwyer, T.J, Geierstanger, B.H, Wemmer, D.E, Schultz, P.G, Spraggon, G, Romesberg, F.E. | | Deposit date: | 2007-04-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs.
J.Am.Chem.Soc., 129, 2007
|
|
7QDX
 
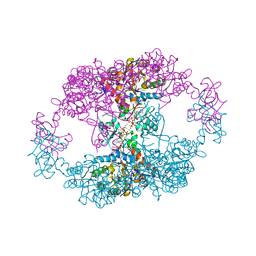 | | bacterial IMPDH chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
1V3O
 
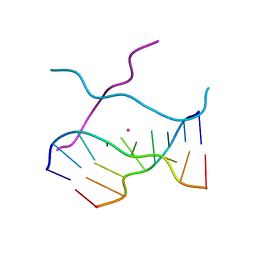 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
8DRL
 
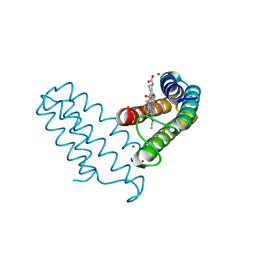 | |
5MPX
 
 | | Crystal structure of Arabidopsis thaliana RNA editing factor MORF1, space group P2(1) | | Descriptor: | Multiple organellar RNA editing factor 1, mitochondrial, SULFATE ION | | Authors: | Haag, S, Schindler, M, Berndt, L, Brennicke, A, Takenaka, M, Weber, G. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9.
Nucleic Acids Res., 45, 2017
|
|
8QQ1
 
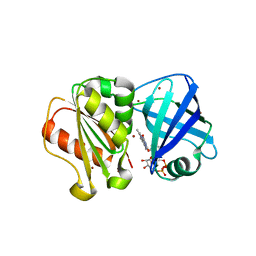 | | SpNOX dehydrogenase domain, mutant F397W in complex with Flavin adenine dinucleotide (FAD) | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Humm, A.S, Dupeux, F, Vermot, A, Petit-Harleim, I, Fieschi, F, Marquez, J.A. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
2PZV
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas Putida (pksi) with bound Phenol | | Descriptor: | PHENOL, Steroid Delta-isomerase | | Authors: | Pybus, B, Caaveiro, J.M.M, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-05-18 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Testing Electrostatic complementarity in Enzyme Catalysis: Hydrogen Bonding in the Ketosteroid Isomerase Oxyanion Hole
PLoS Biol., 4, 2006
|
|
8DRF
 
 | | Zn(II)-bound B2 dimer (H60/H100/H104) | | Descriptor: | HEME C, Soluble cytochrome b562, ZINC ION | | Authors: | Choi, T.S, Tezcan, F.A. | | Deposit date: | 2022-07-20 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of a Flexible, Zn-Selective Protein Scaffold that Displays Anti-Irving-Williams Behavior.
J.Am.Chem.Soc., 144, 2022
|
|
2Q0N
 
 | | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Turnbull, A, Papagrigoriou, E, Pike, A.W, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-22 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide.
To be Published
|
|
5MQY
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile, Cathepsin L1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors.
J. Med. Chem., 60, 2017
|
|
5MR0
 
 | | Thermophilic archaeal branched-chain amino acid transaminases from Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterisation | | Descriptor: | 1,2-ETHANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P, Sayer, C, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the ArchaeaGeoglobus acetivoransandArchaeoglobus fulgidus: Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
8QQ7
 
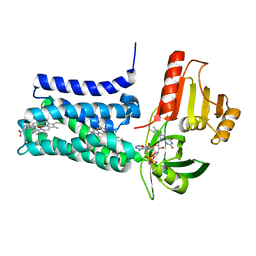 | | Structure of SpNOX: a Bacterial NADPH oxidase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-binding FR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Thepaut, M, Petit-Hartlein, I, Vermot, A, Chaptal, V, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
