8IBS
 
 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E160A/E318A mutant in complex with galactose | | Descriptor: | Beta-galactosidase, alpha-D-galactopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
2P55
 
 | |
2P3B
 
 | | Crystal Structure of the subtype B wild type HIV protease complexed with TL-3 inhibitor | | Descriptor: | benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
5IPQ
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 2 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
1KT9
 
 | | Crystal Structure of C. elegans Ap4A Hydrolase | | Descriptor: | Diadenosine Tetraphosphate Hydrolase | | Authors: | Bailey, S, Sedelnikova, S.E, Blackburn, G.M, Abdelghany, H.M, Baker, P.J, McLennan, A.G, Rafferty, J.B. | | Deposit date: | 2002-01-15 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms
Structure, 10, 2002
|
|
8C29
 
 | | Cryo-EM structure of photosystem II C2S2 supercomplex from Norway spruce (Picea abies) at 2.8 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Kopecny, D, Semchonok, D.A, Kouril, R. | | Deposit date: | 2022-12-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.785 Å) | | Cite: | Cryo-EM structure of a plant photosystem II supercomplex with light-harvesting protein Lhcb8 and alpha-tocopherol.
Nat.Plants, 9, 2023
|
|
1KVV
 
 | | Solution Structure Of Protein SRP19 Of The Archaeoglobus fulgidus Signal Recognition Particle, Minimized Average Structure | | Descriptor: | SRP19 | | Authors: | Pakhomova, O.N, Deep, S, Huang, Q, Zwieb, C, Hinck, A.P. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRP19 of Archaeoglobus fulgidus signal recognition particle.
J.Mol.Biol., 317, 2002
|
|
6ENH
 
 | |
1KO5
 
 | | Crystal structure of gluconate kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Gluconate kinase, MAGNESIUM ION | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
6ES4
 
 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
5IST
 
 | | Staphylococcus aureus Dihydrofolate Reductase complexed with beta-NADPH, cyclic alpha-NADPH anomer and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.723 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
1KQ5
 
 | |
4N4K
 
 | | Kuenenia stuttgartiensis hydroxylamine oxidoreductase soaked in hydroxylamine | | Descriptor: | 1,2-ETHANEDIOL, HEME C, HYDROXYAMINE, ... | | Authors: | Maalcke, W.J, Dietl, A, Marritt, S.J, Butt, J.N, Jetten, M.S.M, Keltjens, J.T, Barends, T.R.M.B, Kartal, B. | | Deposit date: | 2013-10-08 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Biological NO Generation by Octaheme Oxidoreductases.
J.Biol.Chem., 289, 2014
|
|
4MQX
 
 | | CLC-ec1 Fab Complex Cysless A399C-A432C mutant | | Descriptor: | CHLORIDE ION, ERIC, H(+)/Cl(-) exchange transporter ClcA, ... | | Authors: | Basilio, D, Noack, K, Picollo, A, Accardi, A. | | Deposit date: | 2013-09-16 | | Release date: | 2014-04-16 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (3.516 Å) | | Cite: | Conformational changes required for H(+)/Cl(-) exchange mediated by a CLC transporter.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6E7G
 
 | |
4MSD
 
 | | Crystal structure of Schizosaccharomyces pombe AMSH-like protein SST2 T319I mutant | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, AMSH-like protease sst2, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
1KHZ
 
 | | Structure of the ADPR-ase in complex with AMPCPR and Mg | | Descriptor: | ADP-ribose pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2001-12-01 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase.
Biochemistry, 41, 2002
|
|
1KXJ
 
 | | The Crystal Structure of Glutamine Amidotransferase from Thermotoga maritima | | Descriptor: | Amidotransferase hisH, PHOSPHATE ION | | Authors: | Korolev, S, Skarina, T, Evdokimova, E, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Thermotoga maritima
Proteins, 49, 2002
|
|
4N6Q
 
 | | Crystal structure of VosA velvet domain | | Descriptor: | IODIDE ION, NITRATE ION, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
6OQM
 
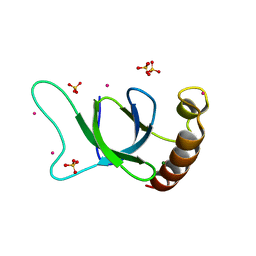 | | crystal structure of the MSH6 PWWP domain | | Descriptor: | DNA mismatch repair protein Msh6, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | crystal structure of the MSH6 PWWP domain
To Be Published
|
|
5ISP
 
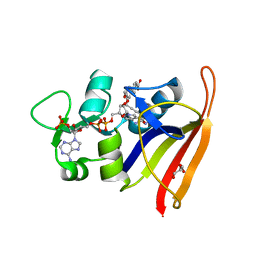 | | Staphylococcus aureus F98Y Dihydrofolate Reductase mutant complexed with beta-NADPH and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
5ISQ
 
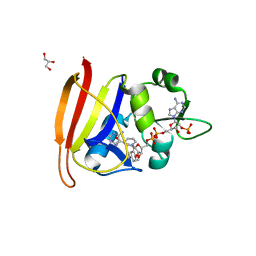 | | Staphylococcus aureus H30N, F98Y Dihydrofolate Reductase mutant complexed with beta-NADPH and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
2P0C
 
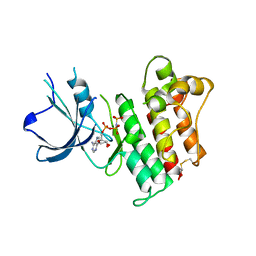 | | Catalytic Domain of the Proto-oncogene Tyrosine-protein Kinase MER | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
1KKH
 
 | | Crystal Structure of the Methanococcus jannaschii Mevalonate Kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate Kinase | | Authors: | Yang, D, Shipman, L.W, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-12-08 | | Release date: | 2002-03-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily.
J.Biol.Chem., 277, 2002
|
|
5I1R
 
 | | Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR and X-ray scattering | | Descriptor: | Nucleocapsid protein p7, ZINC ION | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Quantitative Characterization of Configurational Space Sampled by HIV-1 Nucleocapsid Using Solution NMR, X-ray Scattering and Protein Engineering.
Chemphyschem, 17, 2016
|
|
