6XYN
 
 | | Crystal structure of a proteolytic fragment of NarQ comprising sensor and TM domains | | Descriptor: | NITRATE ION, Nitrate/nitrite sensor histidine kinase NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Gordeliy, V. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a proteolytic fragment of the sensor histidine kinase NarQ
Crystals, 2020
|
|
8VLM
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) E64V-M100W heterodimer | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, ZINC ION | | Authors: | Picard, M.-E, Grenier, G, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
1YBM
 
 | | X-ray structure of selenomethionyl gene product from Arabidopsis thaliana at5g02240 in space group P21212 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, unknown protein At5g02240 | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | X-ray structure of selenomethionyl gene product from Arabidopsis thaliana at5g02240 in space group P21212
To be Published
|
|
2NX2
 
 | | Crystal structure of protein ypsA from Bacillus subtilis, Pfam DUF1273 | | Descriptor: | Hypothetical protein ypsA | | Authors: | Ramagopal, U.A, Alvarado, J, Dickey, M, Reyes, C, Toro, R, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-16 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hypothetical protein ypsA from Bacillus subtilis.
To be Published
|
|
5JD8
 
 | | Crystal structure of the serine endoprotease from Yersinia pestis | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DI(HYDROXYETHYL)ETHER, Periplasmic serine peptidase DegS, ... | | Authors: | Filippova, E.V, Wawrzsak, Z, Sandoval, J, Skarina, T, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the serine endoprotease from Yersinia pestis
To Be Published
|
|
6Z56
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003208 | | Descriptor: | 6-(2-methoxyethoxy)-11-methyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003208
To Be Published
|
|
6Z59
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003816 | | Descriptor: | 11-oxa-8,14,18,19,22-pentazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2,4,6(23),15(22),16,19-heptaen-7-one, SODIUM ION, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003816
To Be Published
|
|
8GBI
 
 | |
8GKX
 
 | |
6Z0D
 
 | |
8G7U
 
 | |
8GL0
 
 | |
8GJ7
 
 | |
6N3Y
 
 | |
8GD8
 
 | |
8GK9
 
 | |
1M7K
 
 | | Solution Structure of the SODD BAG Domain | | Descriptor: | Silencer of Death Domains | | Authors: | Brockmann, C, Leitner, D, Labudde, D, Diehl, A, Sievert, V, Buessow, K, Oschkinat, H. | | Deposit date: | 2002-07-22 | | Release date: | 2002-08-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the SODD BAG domain reveals additional electrostatic interactions in the HSP70 complexes of SODD subfamily BAG domains
Febs Lett., 558, 2004
|
|
2CJQ
 
 | |
6MZC
 
 | | Human TFIID BC core | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
8GIV
 
 | |
8QKV
 
 | | SWR1-nucleosome complex in configuration 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Nucleosome flipping drives kinetic proof reading and processivity of histone exchange by yeast SWR1 complex
To Be Published
|
|
8B7X
 
 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - apo form. | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), SULFATE ION, Siderophore ABC transporter substrate-binding protein | | Authors: | Wilson, K.S, Duhme-Klair, A.K, Blagova, E.V, Bennett, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4YL4
 
 | | 1.1 Angstrom resolution X-ray Crystallographic Structure of Psudoazurin | | Descriptor: | COPPER (II) ION, GLYCEROL, Pseudoazurin | | Authors: | Yamaguchi, T, Asamura, S, Takashina, A, Unno, M, Kohzuma, T. | | Deposit date: | 2015-03-05 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystallographic evidence for the simultaneous presence of axial and rhombic sites in cupredoxins: atomic resolution X-ray crystal structure analysis of pseudoazurin and DFT modelling
Rsc Adv, 6, 2016
|
|
6YG0
 
 | |
8QKU
 
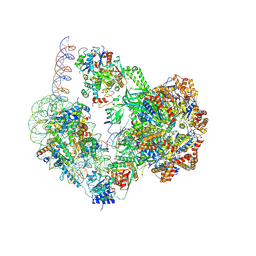 | | SWR1-nucleosome complex in configuration 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Nucleosome flipping drives kinetic proof reading and processivity of histone exchange by yeast SWR1 complex
To Be Published
|
|
