4QVB
 
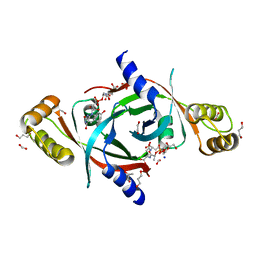 | | Mycobacterium tuberculosis protein Rv1155 in complex with co-enzyme F420 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, COENZYME F420, ... | | Authors: | Mashalidis, E.H, Gittis, A.G, Tomczak, A, Abell, C, Barry III, C.E, Garboczi, D.N. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular insights into the binding of coenzyme F420 to the conserved protein Rv1155 from Mycobacterium tuberculosis.
Protein Sci., 24, 2015
|
|
1THA
 
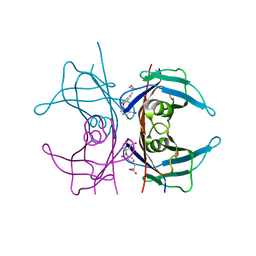 | | MECHANISM OF MOLECULAR RECOGNITION. STRUCTURAL ASPECTS OF 3,3'-DIIODO-L-THYRONINE BINDING TO HUMAN SERUM TRANSTHYRETIN | | Descriptor: | 3,3'-DEIODO-THYROXINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Luft, J, Cody, V. | | Deposit date: | 1991-11-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of molecular recognition. Structural aspects of 3,3'-diiodo-L-thyronine binding to human serum transthyretin.
J.Biol.Chem., 267, 1992
|
|
3X2N
 
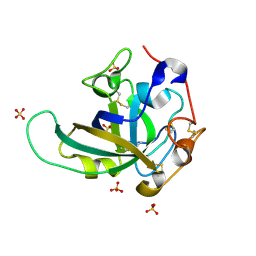 | | Proton relay pathway in inverting cellulase | | Descriptor: | Endoglucanase V-like protein, SULFATE ION | | Authors: | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
2IGQ
 
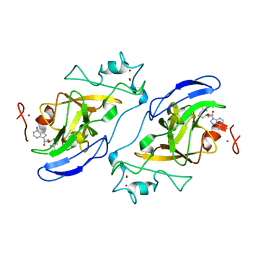 | | Human euchromatic histone methyltransferase 1 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, euchromatic histone methyltransferase 1 | | Authors: | Min, J, Wu, H, Antoshenko, T, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-23 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
5KCT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KD5
 
 | |
5KF9
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Holden, H.M, Tipton, P.A. | | Deposit date: | 2016-06-12 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
6W3T
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-norvaline | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, NORVALINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
3EQB
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | 2-Alkylamino- and alkoxy-substituted 2-amino-1,3,4-oxadiazoles-O-Alkyl benzohydroxamate esters replacements retain the desired inhibition and selectivity against MEK (MAP ERK kinase).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4RBP
 
 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
5KGJ
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with galactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-alpha-D-galactopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
4IJD
 
 | | Crystal structure of methyltransferase domain of human PR domain-containing protein 9 | | Descriptor: | Histone-lysine N-methyltransferase PRDM9, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Dombrovski, L, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of methyltransferase domain of human PR domain-containing protein 9
To be Published
|
|
6VQ2
 
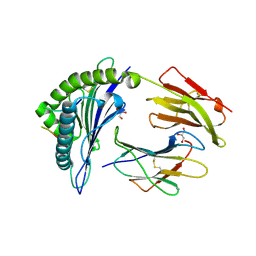 | | HLA-B*27:05 presenting an HIV-1 14mer peptide | | Descriptor: | 14-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQD
 
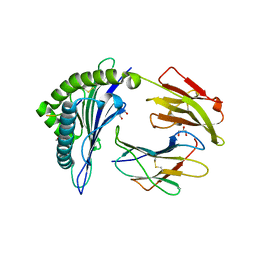 | | HLA-B*27:05 presenting an HIV-1 8mer peptide | | Descriptor: | 8-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQE
 
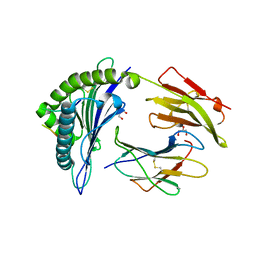 | | HLA-B*27:05 presenting an HIV-1 13mer peptide | | Descriptor: | 13-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
3EOZ
 
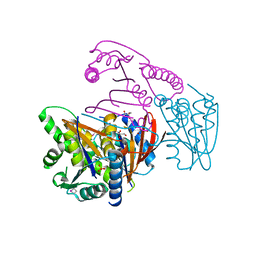 | | Crystal Structure of Phosphoglycerate Mutase from Plasmodium Falciparum, PFD0660w | | Descriptor: | GLYCEROL, PHOSPHATE ION, putative Phosphoglycerate mutase | | Authors: | Wernimont, A.K, Tempel, W, Lam, A, Zhao, Y, Lew, J, Lin, Y.H, Wasney, G, Vedadi, M, Kozieradzki, I, Cossar, D, Schapira, M, Weigelt, J, Arrowsmith, C.H, Bochkarev, A, Edwards, A.M, Hui, R, Pizarro, J, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a new phosphatase from Plasmodium.
Mol.Biochem.Parasitol., 179, 2011
|
|
7AN6
 
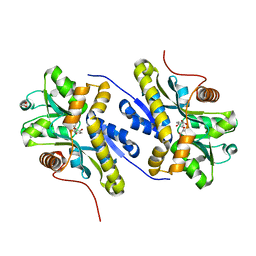 | | Enzyme of biosynthetic pathway | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-10-11 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
6VPZ
 
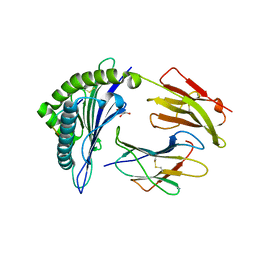 | | HLA-B*27:05 presenting an HIV-1 11mer peptide | | Descriptor: | 11-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
5KGA
 
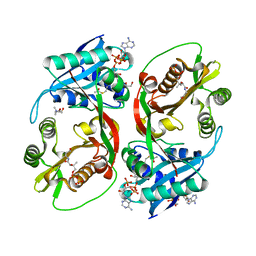 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant D287N, in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
2W9Y
 
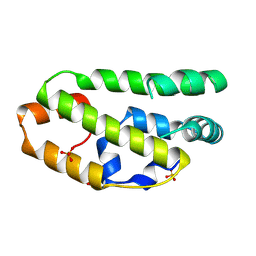 | | The structure of the lipid binding protein Ce-FAR-7 from Caenorhabditis elegans | | Descriptor: | FATTY ACID/RETINOL BINDING PROTEIN PROTEIN 7, ISOFORM A, CONFIRMED BY TRANSCRIPT EVIDENCE, ... | | Authors: | Jordanova, R, Groves, M.R, Tucker, P.A. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-20 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fatty Acid and Retinoid Binding Proteins Have Distinct Binding Pockets for the Two Types of Cargo
J.Biol.Chem., 284, 2009
|
|
7AMQ
 
 | | Crystal structure of the complex of HuJovi-1 Fab with the human TRBC2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Human A6 T-cell receptor alpha chain, ... | | Authors: | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | Deposit date: | 2020-10-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
6VR0
 
 | | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Liu, D, Ballicora, M, Iglesias, A, Asencion, M, Figueroa, C. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A
To Be Published
|
|
6Q0W
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to Indisulam and RBM39 | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
7AMR
 
 | | Crystal structure of the complex of the KFN mutant of Jovi-1 Fab with human TRBC1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Human Jovi-1 Fab fragment, ... | | Authors: | Ferrari, M, Bulek, A, Bughda, R, Jha, R, Welin, M, Logan, D.T, Sewell, A, Onuoha, S, Pule, M. | | Deposit date: | 2020-10-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Res Sq, 2022
|
|
5JYT
 
 | |
