6PHX
 
 | |
2VXU
 
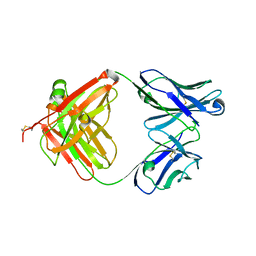 | | Crystal structure of murine reference antibody 125-2H Fab fragment | | 分子名称: | MURINE IGG 125-2H | | 著者 | Argiriadi, M.A, Xiang, T, Wu, C, Ghayur, T, Borhani, D.W. | | 登録日 | 2008-07-10 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Unusual Water-Mediated Antigenic Recognition of the Proinflammatory Cytokine Interleukin-18.
J.Biol.Chem., 284, 2009
|
|
5KGH
 
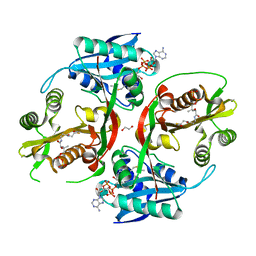 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant Y297F | | 分子名称: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | 著者 | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | 登録日 | 2016-06-13 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
7ZLP
 
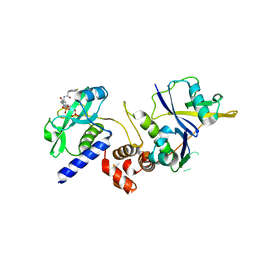 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 9 | | 分子名称: | Elongin-B, Elongin-C, PHOSPHATE ION, ... | | 著者 | Ramachandran, S, Ciulli, A, Makukhin, N. | | 登録日 | 2022-04-15 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
1JW4
 
 | |
4R5N
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | 分子名称: | (4R,4a'S,10a'R)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | 著者 | Vigers, G.P.A, Smith, D. | | 登録日 | 2014-08-21 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
7ZKE
 
 | | Mot1:TBP:DNA - pre-hydrolysis state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (36-MER), ... | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6PKU
 
 | | Guinea pig N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA) catalytic domain (C51S C221S) in complex with N-acetyl-alpha-D-glucosamine (alpha-GlcNAc) and mannose 6-phosphate (M6P) | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-alpha-D-mannopyranose, ... | | 著者 | Gorelik, A, Illes, K, Nagar, B. | | 登録日 | 2019-06-29 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Structure, 28, 2020
|
|
1T0M
 
 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | 分子名称: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | 著者 | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | 登録日 | 2004-04-12 | | 公開日 | 2004-11-23 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
3E8E
 
 | | Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors | | 分子名称: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, PKI inhibitor peptide, cAMP-dependent protein kinase catalytic subunit alpha | | 著者 | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | 登録日 | 2008-08-19 | | 公開日 | 2008-11-18 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5K9Y
 
 | | Crystal structure of a thermophilic xylanase A from Bacillus subtilis 1A1 quadruple mutant Q7H/G13R/S22P/S179C | | 分子名称: | Endo-1,4-beta-xylanase A | | 著者 | Pinheiro, M.P, Ferreira, T.L, Silva, S.R.B, Fuzo, C.A, Silva, S.R, Lourenzoni, M.R, Vieira, D.S, Ward, R.J, Nonato, M.C. | | 登録日 | 2016-06-01 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The role of local residue environmental changes in thermostable mutants of the GH11 xylanase from Bacillus subtilis.
Int. J. Biol. Macromol., 97, 2017
|
|
5KA6
 
 | |
4RBR
 
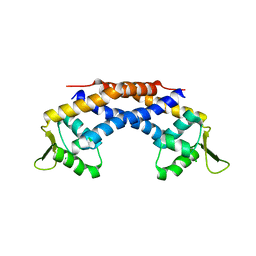 | | Crystal structure of Repressor of Toxin (Rot), a central regulator of Staphylococcus aureus virulence | | 分子名称: | CHLORIDE ION, HTH-type transcriptional regulator rot | | 著者 | Killikelly, A, Jakoncic, J, Sampson, J.M, Kong, X.-P. | | 登録日 | 2014-09-12 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Based Functional Characterization of Repressor of Toxin (Rot), a Central Regulator of Staphylococcus aureus Virulence.
J.Bacteriol., 197, 2015
|
|
7ZK8
 
 | | ABCB1 L971C mutant (mABCB1) in the outward facing state bound to AAC | | 分子名称: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | 著者 | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
3ZOL
 
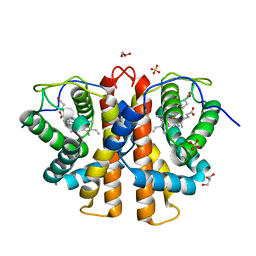 | | M.acetivorans protoglobin F93Y mutant in complex with cyanide | | 分子名称: | CYANIDE ION, GLYCEROL, PROTOGLOBIN, ... | | 著者 | Tilleman, L, Abbruzzetti, S, Ciaccio, C, De Sanctis, G, Nardini, M, Pesce, A, Desmet, F, Moens, L, Van Doorslaer, S, Bruno, S, Bolognesi, M, Ascenzi, P, Coletta, M, Viappiani, C, Dewilde, S. | | 登録日 | 2013-02-22 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Bases for the Regulation of Co Binding in the Archaeal Protoglobin from Methanosarcina Acetivorans.
Plos One, 10, 2015
|
|
7ZLR
 
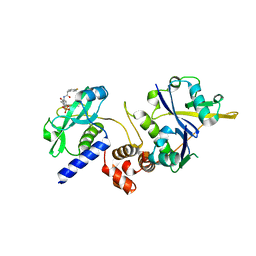 | | Crystal structure of SOCS2:ElonginB:ElonginC in complex with compound 13 | | 分子名称: | Elongin-B, Elongin-C, Suppressor of cytokine signaling 2, ... | | 著者 | Ramachandran, S, Ciulli, A, Makukhin, N. | | 登録日 | 2022-04-15 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Nat Commun, 14, 2023
|
|
3X2L
 
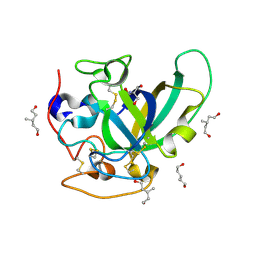 | | X-ray structure of PcCel45A apo form at 95K. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | 著者 | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (0.83 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
1T64
 
 | | Crystal Structure of human HDAC8 complexed with Trichostatin A | | 分子名称: | CALCIUM ION, Histone deacetylase 8, SODIUM ION, ... | | 著者 | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | 登録日 | 2004-05-05 | | 公開日 | 2004-07-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
8A4J
 
 | | Human GDAP1, A247V mutant | | 分子名称: | Ganglioside-induced differentiation-associated protein 1 | | 著者 | Sutinen, A, Kursula, P. | | 登録日 | 2022-06-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Conserved intramolecular networks in GDAP1 are closely connected to CMT-linked mutations and protein stability.
Plos One, 18, 2023
|
|
6W5K
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5g | | 分子名称: | 3C-LIKE PROTEASE, N~2~-{[2-(3-chlorophenyl)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
8A4K
 
 | | Human GDAP1, R282H mutant | | 分子名称: | Ganglioside-induced differentiation-associated protein 1 | | 著者 | Sutinen, A, Kursula, P. | | 登録日 | 2022-06-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Conserved intramolecular networks in GDAP1 are closely connected to CMT-linked mutations and protein stability.
Plos One, 18, 2023
|
|
2IEN
 
 | | Crystal structure analysis of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-09-19 | | 公開日 | 2006-10-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
5KH9
 
 | | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | 分子名称: | 6-methyl-5-[(4-propan-2-ylphenyl)amino]-2~{H}-1,2,4-triazin-3-one, FORMIC ACID, Histone deacetylase 6, ... | | 著者 | Harding, R.J, Tempel, W, Ravichandran, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | 登録日 | 2016-06-14 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
7B3S
 
 | | OXA-10 beta-lactamase with S67Dha modification | | 分子名称: | Beta-lactamase OXA-10, CARBON DIOXIDE, SODIUM ION, ... | | 著者 | Lang, P.A, Brem, J, Schofield, C.J. | | 登録日 | 2020-12-01 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Studies on enmetazobactam clarify mechanisms of widely used beta-lactamase inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KI7
 
 | | Structural impact of single ribonucleotides in DNA | | 分子名称: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*GP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*CP*C)-R(P*G)-D(P*GP*TP*AP*G)-3') | | 著者 | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | 登録日 | 2016-06-16 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
