4YPT
 
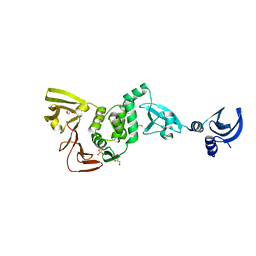 | | X-ray structural of three tandemly linked domains of nsp3 from murine hepatitis virus at 2.60 Angstroms resolution | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Chen, Y, Savinov, S.N, Mielech, A.M, Cao, T, Baker, S.C, Mesecar, A.D. | | Deposit date: | 2015-03-13 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6009 Å) | | Cite: | X-ray Structural and Functional Studies of the Three Tandemly Linked Domains of Non-structural Protein 3 (nsp3) from Murine Hepatitis Virus Reveal Conserved Functions.
J.Biol.Chem., 290, 2015
|
|
6XK0
 
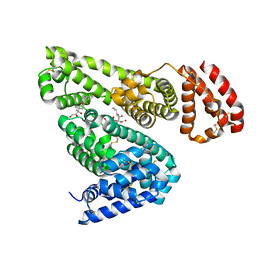 | | Albumin-dexamethasone complex | | Descriptor: | Albumin, CITRATE ANION, DEXAMETHASONE, ... | | Authors: | Czub, M.P, Majorek, K.A, Shabalin, I.G, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2020-06-24 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular determinants of vascular transport of dexamethasone in COVID-19 therapy.
Iucrj, 7, 2020
|
|
4YQ0
 
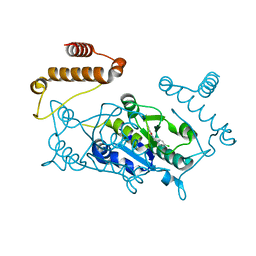 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | 4-amino-N-(4-chlorobenzyl)-1,2,5-oxadiazole-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To be published
|
|
5HJR
 
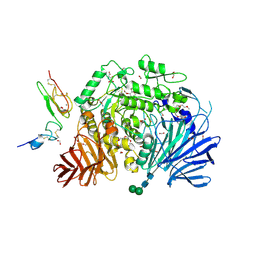 | | Murine endoplasmic reticulum alpha-glucosidase II with bound covalent intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoro-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-01-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7LDX
 
 | | SARS-CoV-2 Main protease immature form - F2X Entry Library E06 fragment | | Descriptor: | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
6G1M
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open and closed form | | Descriptor: | Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Beloti, L, Frese, A, Mayol, O, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
7LFP
 
 | | SARS-CoV-2 Main protease immature form - F2X Entry Library G05 fragment | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
8CYE
 
 | |
6G2F
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 16 | | Descriptor: | 4-[5-(7-fluoranylquinolin-4-yl)-1-methyl-imidazol-4-yl]-3,5-dimethyl-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
8OUS
 
 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 19 | | Descriptor: | (1S)-1-[4-[6-azanyl-5-(trifluoromethyloxy)pyridin-3-yl]-1-(3-morpholin-4-yl-1-bicyclo[1.1.1]pentanyl)imidazol-2-yl]-2-methyl-propan-1-ol, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, McEwan, P.A, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7R71
 
 | | Crystal Structure of the UbArk2C-UbcH5b~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin,E3 ubiquitin-protein ligase RNF165, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
6SXR
 
 | | E221Q mutant of GH54 a-l-arabinofuranosidase soaked with 4-nitrophenyl a-l-arabinofuranoside | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Davies, G.J, Nin-Hill, A, Rovira, C. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
8Q5Q
 
 | | d(ATTTC)3 dimeric structure | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*(DNR)P*AP*TP*TP*TP*CP*AP*TP*TP*TP*C)-3') | | Authors: | Trajkovski, M, Pastore, A, Plavec, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Dimeric structures of DNA ATTTC repeats promoted by divalent cations.
Nucleic Acids Res., 52, 2024
|
|
5O5A
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ032 | | Descriptor: | 1-ethyl-~{N}-methyl-2,3-bis(oxidanylidene)-~{N}-[(1-phenylpyrazol-4-yl)methyl]-4~{H}-quinoxaline-6-carboxamide, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
6FYZ
 
 | | Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor | | Descriptor: | (2~{S})-2-(2-fluorophenyl)-2-[4-(2-methylpyrimidin-5-yl)phenyl]-~{N}-oxidanyl-ethanamide, Histone deacetylase 4, SODIUM ION, ... | | Authors: | Luckhurst, C.A, Maillard, M.C, Dominguez, C. | | Deposit date: | 2018-03-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5NYN
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in complex with gluathione | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5Z84
 
 | | The structure of azide-bound cytochrome c oxidase determined using the crystals exposed to 20 mM azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
8OUR
 
 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 5-[1-(3-morpholin-4-yl-1-bicyclo[1.1.1]pentanyl)-2-propan-2-yl-imidazol-4-yl]-3-(trifluoromethyloxy)pyridin-2-amine, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | Zebisch, M, McEwan, P.A, Barker, J.J, Cross, J.B. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
3NM3
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, THIAMIN PHOSPHATE, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
7LFE
 
 | | SARS-CoV-2 Main protease immature form - F2X Entry Library E03 fragment | | Descriptor: | (2R,4R)-1-phenylhexahydropyrimidine-2,4-diol, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
6QSD
 
 | | Crystal structure of Pizza6S | | Descriptor: | Pizza6S, SULFATE ION | | Authors: | Noguchi, H, Vandebroek, L, Kamata, K, Tame, J.R.H, Van Meervelt, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Hybrid assemblies of a symmetric designer protein and polyoxometalates with matching symmetry.
Chem.Commun.(Camb.), 56, 2020
|
|
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
7LG7
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain complex with PARG345 | | Descriptor: | 3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]-N-{[2-(morpholin-4-yl)ethyl]sulfonyl}propanamide, Non-structural protein 3, SULFATE ION | | Authors: | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2021-01-19 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6MR6
 
 | |
