6FPU
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex with 6-amino-2-((((3aS,5aR,8bS)-2,2,7,7-tetramethyltetrahydro-3aH-bis([1,3]dioxolo)[4,5-b:4',5'-d]pyran-3a-yl)methyl)amino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-azanyl-2-[[(1~{R},2~{S},6~{S},9~{R})-4,4,11,11-tetramethyl-3,5,7,10,12-pentaoxatricyclo[7.3.0.0^{2,6}]dodecan-6-yl]methylamino]-3,7-dihydroimidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2018-02-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Sugar Acetonides are a Superior Motif for Addressing the Large, Solvent-Exposed Ribose-33 Pocket of tRNA-Guanine Transglycosylase.
Chemistry, 24, 2018
|
|
7O4C
 
 | |
4WK2
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CB3
 
 | |
8OZQ
 
 | | In situ subtomogram average of Prototype Foamy Virus Env hexamer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 2024
|
|
8TBT
 
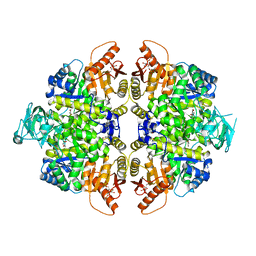 | | Structure of human erythrocyte pyruvate kinase in complex with an allosteric activator Compound 2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Jin, L, Padyana, A. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-Based Design of AG-946, a Pyruvate Kinase Activator.
Chemmedchem, 19, 2024
|
|
6YG6
 
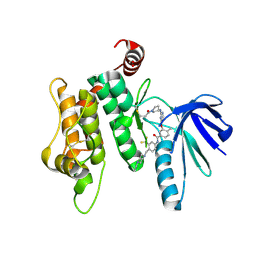 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor TL10-105 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{S})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
7SNU
 
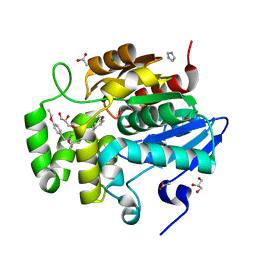 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | Descriptor: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
8CM3
 
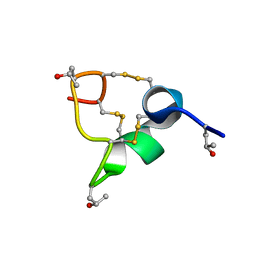 | |
6YHD
 
 | | tRNA-guanine Transglycosylase (TGT) labeled with 5-fluorotryptophan in co-crystallized complex with 6-amino-4-(2-((2R,3S,4R,5R)-3,4-dihydroxy-5-methoxytetrahydrofuran-2-yl)ethyl)-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Co-crystallization, nanoESI-MS and 19F NMR reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
6N5G
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6CC3
 
 | |
6FPP
 
 | | Structure of S. pombe Mmi1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
6SKE
 
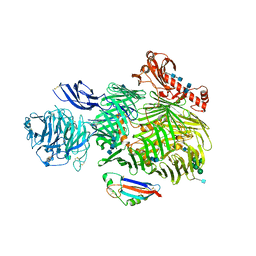 | | Teneurin 2 in complex with Latrophilin 2 Lec domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor L2, ... | | Authors: | Shahin, M, Jackson, V.A, Carrasquero, M, Lowe, E, Seiradake, E. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural Basis of Teneurin-Latrophilin Interaction in Repulsive Guidance of Migrating Neurons.
Cell, 180, 2020
|
|
8PIP
 
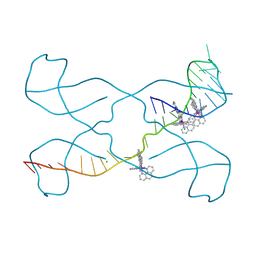 | |
8TWD
 
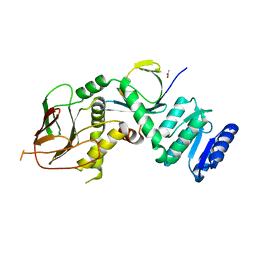 | | Structure of IraM-bound RssB | | Descriptor: | ACETATE ION, Anti-adapter protein IraM, Regulator of RpoS | | Authors: | Brugger, C, Deaconescu, A.M. | | Deposit date: | 2023-08-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | IraM remodels the RssB segmented helical linker to stabilize sigma s against degradation by ClpXP.
J.Biol.Chem., 300, 2023
|
|
8OZJ
 
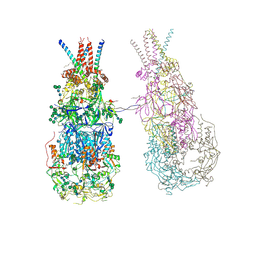 | | In situ cryoEM structure of Prototype Foamy Virus Env dimer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 2024
|
|
6N6T
 
 | | OXA-23 mutant F110A/M221A low pH form | | Descriptor: | Beta-lactamase oxa23, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Role of the Hydrophobic Bridge in the Carbapenemase Activity of Class D beta-Lactamases.
Antimicrob. Agents Chemother., 63, 2019
|
|
8CO5
 
 | |
4WPL
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form I | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8OZP
 
 | | In situ subtomogram average of Prototype Foamy Virus Env pentamer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 2024
|
|
5J5Z
 
 | | Crystal structure of the D444V disease-causing mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Mizsei, R, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2016-04-04 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase.
Free Radic. Biol. Med., 124, 2018
|
|
7ZNF
 
 | |
6FT3
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{R})-cyclopropyl(oxidanyl)methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
4WQ8
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - tetrathionate soak | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.3989 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
