6QKF
 
 | |
1K4G
 
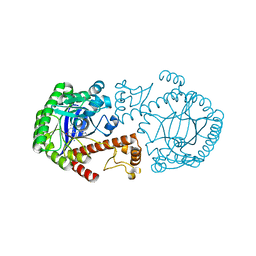 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
ChemBioChem, 3, 2002
|
|
4V1I
 
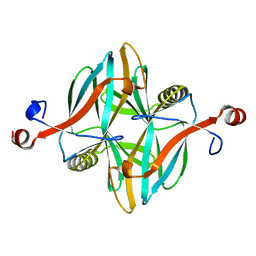 | | Structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 at medium resolution | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6Z5C
 
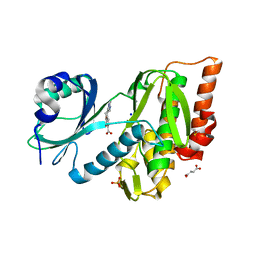 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004070 | | Descriptor: | 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004070
To Be Published
|
|
5X9G
 
 | |
6ZFK
 
 | |
4CZN
 
 | |
2D4X
 
 | |
8Q4G
 
 | | Thin filament from FIB milled relaxed left ventricular mouse myofibrils | | Descriptor: | Actin, alpha cardiac muscle 1, Tropomyosin alpha-1 chain | | Authors: | Tamborrini, D, Wang, Z, Wagner, T, Tacke, S, Stabrin, M, Grange, M, Kho, A.L, Bennet, P, Rees, M, Gautel, M, Raunser, S. | | Deposit date: | 2023-08-06 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of the native myosin filament in the relaxed cardiac sarcomere.
Nature, 623, 2023
|
|
5IU4
 
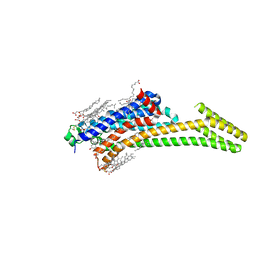 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with ZM241385 at 1.7A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
1TPM
 
 | | SOLUTION STRUCTURE OF THE FIBRIN BINDING FINGER DOMAIN OF TISSUE-TYPE PLASMINOGEN ACTIVATOR DETERMINED BY 1H NUCLEAR MAGNETIC RESONANCE | | Descriptor: | TISSUE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Downing, A.K, Driscoll, P.C, Harvey, T.S, Dudgeon, T.J, Smith, B.O, Baron, M, Campbell, I.D. | | Deposit date: | 1993-05-26 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance.
J.Mol.Biol., 225, 1992
|
|
6EL3
 
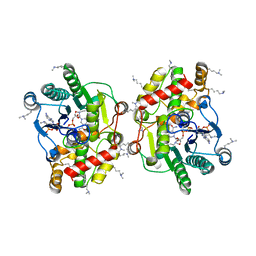 | | Structure of Progesterone 5beta-Reductase from Arabidopsis thaliana in complex with NADP | | Descriptor: | 3-oxo-Delta(4,5)-steroid 5-beta-reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Muller, Y.A, Schmidt, K, Egerer-Sieber, C. | | Deposit date: | 2017-09-27 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | PRISEs (progesterone 5 beta-reductase and/or iridoid synthase-like 1,4-enone reductases): Catalytic and substrate promiscuity allows for realization of multiple pathways in plant metabolism.
Phytochemistry, 156, 2018
|
|
2OQI
 
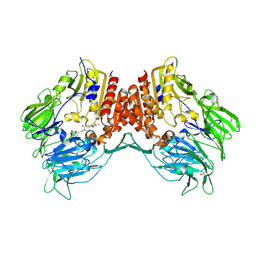 | | Human Dipeptidyl Peptidase IV (DPP4) with Piperidinone-constrained phenethylamine | | Descriptor: | (4R,5R)-5-AMINO-1-[2-(1,3-BENZODIOXOL-5-YL)ETHYL]-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-2-ONE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D, Backes, B.J, Lai, C, Lubben, T.H, Ballaron, S.J, Beno, D.W, Kempf-Grote, A.J, Sham, H.L, Trevillyan, J.M. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
5XBP
 
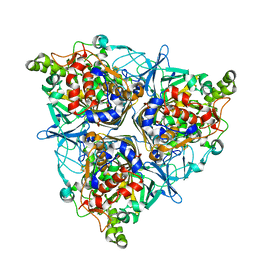 | | Oxygenase component of 3-nitrotoluene dioxygenase from Diaphorobacter sp. strain DS2 | | Descriptor: | 3NT oxygenase alpha subunit, 3NT oxygenase beta subunit, FE (III) ION, ... | | Authors: | Ramaswamy, S, Kumari, A, Singh, D, Gurunath, R. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional studies of ferredoxin and oxygenase components of 3-nitrotoluene dioxygenase from Diaphorobacter sp. strain DS2.
PLoS ONE, 12, 2017
|
|
6ZFJ
 
 | |
2DC0
 
 | | Crystal structure of amidase | | Descriptor: | probable amidase | | Authors: | Ohshima, T, Sakuraba, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Satoh, S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-17 | | Release date: | 2007-01-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amidase
To be Published
|
|
3AE3
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Nitro-N-phenyl-benzamide | | Descriptor: | 2-nitro-N-phenylbenzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with 2-Nitro-N-phenyl-benzamide
To be Published
|
|
6ZGB
 
 | | glutamate transporter homologue Glttk in complex with a photo cage compound | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[(2-nitrophenyl)methoxy]butanedioic acid, DI(HYDROXYETHYL)ETHER, Proton/glutamate symporter, ... | | Authors: | Arkhipova, V, Slotboom, D.J, Guskov, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Aspects of Photopharmacology: Insight into the Binding of Photoswitchable and Photocaged Inhibitors to the Glutamate Transporter Homologue.
J.Am.Chem.Soc., 143, 2021
|
|
3AFN
 
 | | Crystal structure of aldose reductase A1-R complexed with NADP | | Descriptor: | Carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TERTIARY-BUTYL ALCOHOL | | Authors: | Takase, R, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-03-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1
Biochim.Biophys.Acta, 1804, 2010
|
|
1U24
 
 | | Crystal structure of Selenomonas ruminantium phytase | | Descriptor: | myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
6ZJ9
 
 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
5XGY
 
 | | Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Shokeen, A, Sharma, P, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-04-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution
To Be Published
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6ZGF
 
 | | Spike Protein of RaTG13 Bat Coronavirus in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4LWV
 
 | | The 2.3A Crystal Structure of Humanized Xenopus MDM2 with RO5545353 | | Descriptor: | (2S,3R,4R,5R)-N-(4-carbamoyl-2-methoxyphenyl)-2'-chloro-4-(3-chloro-2-fluorophenyl)-2-(2,2-dimethylpropyl)-5'-oxo-4',5'-dihydrospiro[pyrrolidine-3,6'-thieno[3,2-b]pyrrole]-5-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C, Janson, C.A. | | Deposit date: | 2013-07-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of Potent and Orally Active p53-MDM2 Inhibitors RO5353 and RO2468 for Potential Clinical Development.
ACS MED.CHEM.LETT., 5, 2014
|
|
