6XKU
 
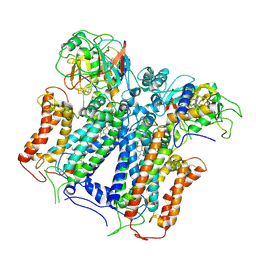 | | R. capsulatus cyt bc1 with one FeS protein in b position and one in c position (CIII2 b-c) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
8PQY
 
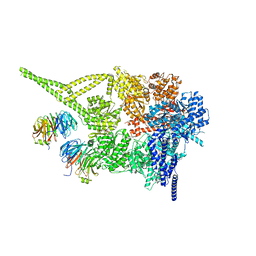 | | Cytoplasmic dynein-1 motor domain bound to LIS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
7A6W
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 33-(Z) | | Descriptor: | (2S,9S,12R,20Z)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-28,31-dimethoxy-11,18,23,26-tetraoxa-4-azatetracyclo[25.2.2.113,17.04,9]dotriaconta-1(29),13(32),14,16,20,27,30-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7OWE
 
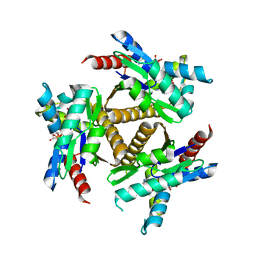 | | Odinarchaeota Adenylate kinase (OdinAK) in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Grundstrom, C, Aberg-Zingmark, E, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
8PR4
 
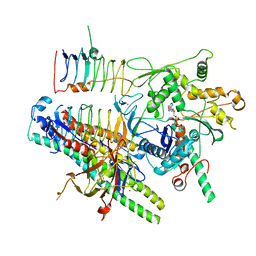 | | Dynactin pointed end bound to JIP3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Arp11, C-Jun-amino-terminal kinase-interacting protein 3, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
7OWL
 
 | | Odinarchaeota Adenylate kinase (OdinAK) in complex with CTP | | Descriptor: | Adenylate kinase, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
7OWK
 
 | | Odinarchaeota Adenylate kinase (OdinAK) in complex with dTTP | | Descriptor: | Adenylate kinase, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
7OWH
 
 | | Odinarchaeota Adenylate kinase (OdinAK) native structure | | Descriptor: | Adenylate kinase, CHLORIDE ION | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
6ELE
 
 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, fAB heavy chain, ... | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6XQU
 
 | |
7OWJ
 
 | | Odinarchaeota Adenylate kinase (OdinAK) in complex with GTP | | Descriptor: | Adenylate kinase, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Aberg-Zingmark, E, Grundstrom, C, Verma, A, Wolf-Watz, M, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2021-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Sci Adv, 8, 2022
|
|
7P2N
 
 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
8FAY
 
 | | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*CP*AP*(NR1)P*GP*TP*CP*T)-3'), ... | | Authors: | Trasvina-Arenas, C.H, Lin, W.J, Demir, M, Fisher, A.J, David, S.S, Horvath, M.P. | | Deposit date: | 2022-11-29 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Human MUTYH adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G)
To Be Published
|
|
7P55
 
 | |
7OFC
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
6U0B
 
 | |
6XWZ
 
 | |
5SU5
 
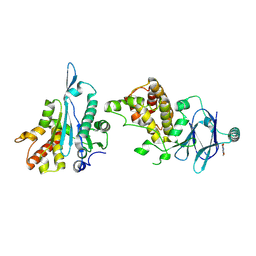 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03E05 from the F2X-Universal Library | | Descriptor: | 5-bromo-1-methylpyrimidine-2,4(1H,3H)-dione, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7P2K
 
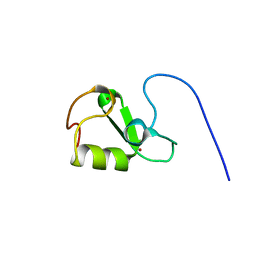 | | Solution NMR Structure of Arginine to Cysteine mutant of Arkadia RING domain. | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Raptis, V, Marousis, K.D, Birkou, M, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Impact of a Single Nucleotide Polymorphism on the 3D Protein Structure and Ubiquitination Activity of E3 Ubiquitin Ligase Arkadia.
Front Mol Biosci, 9, 2022
|
|
8PAW
 
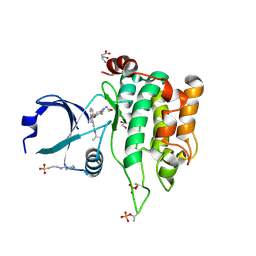 | | Crystal structure of MST1 with a MAP4K1 SMOL inhibitor | | Descriptor: | 1-[3,5-bis(fluoranyl)-4-[[3-(1-propan-2-ylpyrazol-3-yl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-3-(2-methoxyethyl)urea, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ASPARTIC ACID, ... | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
5STN
 
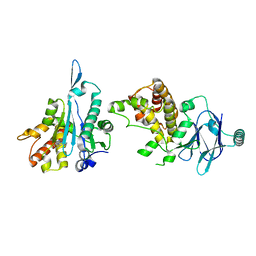 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03A11 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-methyl-1-{(1S,2R)-2-[(1,2,4-triazolidin-1-yl)methyl]cyclohexyl}methanamine, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7PKO
 
 | | CryoEM structure of Rotavirus NSP2 | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PKP
 
 | | NSP2 RNP complex | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8PAS
 
 | | Crystal structure of MAP4K1 with a SMOL inhibitor | | Descriptor: | 4-[2,6-bis(fluoranyl)-4-(3-morpholin-4-ylpropylcarbamoylamino)phenoxy]-~{N}-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]-1~{H}-pyrrolo[2,3-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8AC6
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): medium class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
