7Q53
 
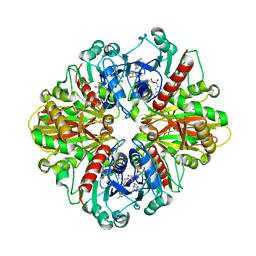 | | Single Particle Cryo-EM structure of photosynthetic A2B2 glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PV7
 
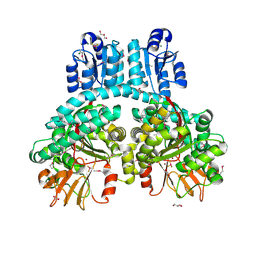 | | Crystal structure of dimeric Porphyromonas gingivalis PorX, a type 9 secretion system response regulator. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Schmitz, C.A, Madej, M, Potempa, J, Sola, M. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors.
Nucleic Acids Res., 50, 2022
|
|
7PWT
 
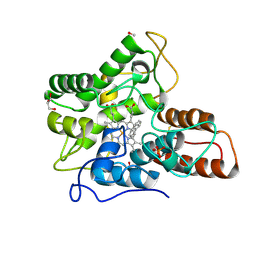
 | | Crystal structure of 14-3-3 sigma in complex with a C-terminal Estrogen Receptor alpha phosphopeptide, stabilised by pyrrolidone derivative 228 | | Descriptor: | (2~{R})-1-(2-hydroxyphenyl)-2-(4-nitrophenyl)-4-oxidanyl-3-(phenylcarbonyl)-2~{H}-pyrrol-5-one, 14-3-3 protein sigma, C-terminus of Estrogen receptor alpha, ... | | Authors: | Andrei, S.A, Bosica, F, O'Mahony, G, Ottmann, C. | | Deposit date: | 2021-10-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Designing Selective Drug-like Molecular Glues for the Glucocorticoid Receptor/14-3-3 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7PN4
 
 | |
7PQH
 
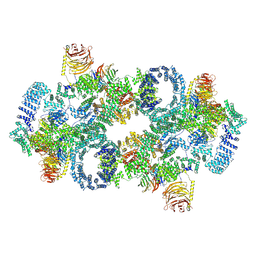 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PN5
 
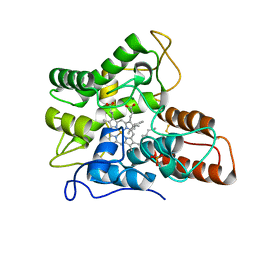 | |
7PN7
 
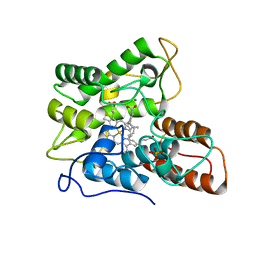 | |
7PN8
 
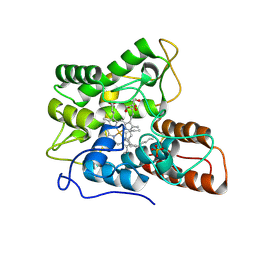 | |
7PN6
 
 | |
7PN9
 
 | |
7PNA
 
 | |
7Q4W
 
 | | CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kumar, A, Saura, P, Poeverlein, M.C, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
7PSC
 
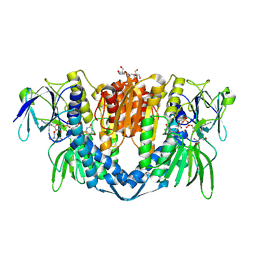 | | Crystal structure of the disease-causing I358T mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Nemes-Nikodem, E, Szabo, E, Zambo, Z, Vass, K.R, Taberman, H, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2021-09-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Structural and Biochemical Investigation of Selected Pathogenic Mutants of the Human Dihydrolipoamide Dehydrogenase.
Int J Mol Sci, 24, 2023
|
|
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | Descriptor: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | Deposit date: | 2021-11-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
6PIW
 
 | |
7AN1
 
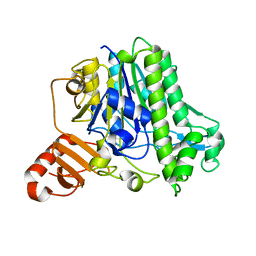 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1636-S1_20) | | Descriptor: | Arylsulfatase, CALCIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
6PJ1
 
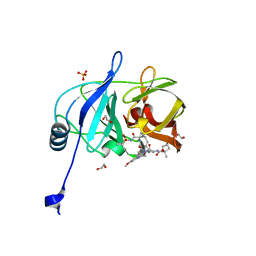 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-4(AJ-74) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
7ANB
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
6P1P
 
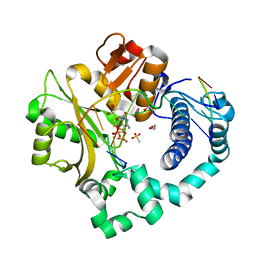 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and bound incoming dCMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
6P1V
 
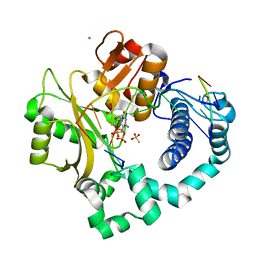 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing undamaged template dG and bound incoming dCMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
6PKH
 
 | |
7PTI
 
 | |
8CCX
 
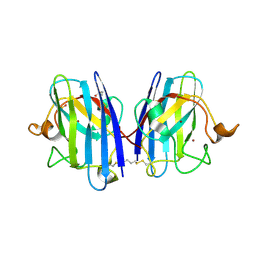 | | Human SOD1 in complex with S-XL6 cross-linker | | Descriptor: | COPPER (II) ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Hossain, A, Agar, J.N, Hasnain, S.S. | | Deposit date: | 2023-01-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Evaluating protein cross-linking as a therapeutic strategy to stabilize SOD1 variants in a mouse model of familial ALS.
Plos Biol., 22, 2024
|
|
6PLD
 
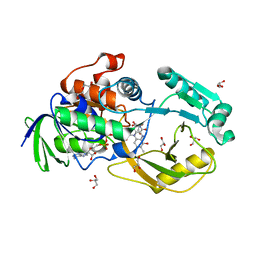 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with 6-OH-FAD - Green fraction | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | Deposit date: | 2019-06-30 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
8ELJ
 
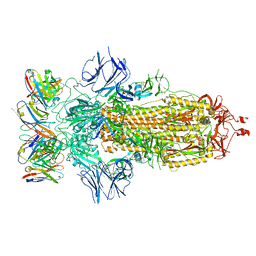 | | SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ICO-hu23 antibody Fab heavy chain, ... | | Authors: | Yee, A.W, Morizumi, T, Kim, K, Kuo, A, Ernst, O.P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
