1VZX
 
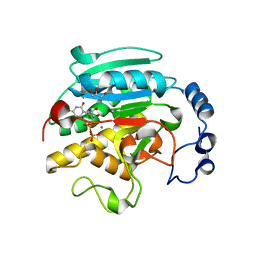 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-28 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
5VTM
 
 | |
6DS6
 
 | |
4OIP
 
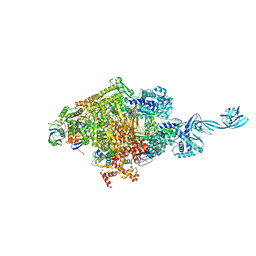 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIR
 
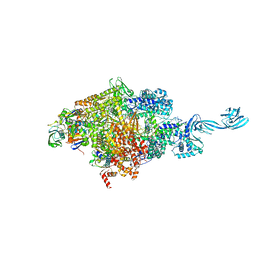 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4FD3
 
 | |
6XNX
 
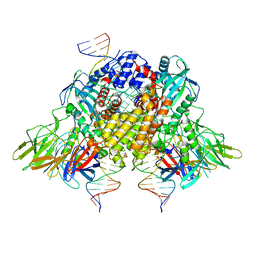 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Dynamic-Form) | | Descriptor: | 12RSS integration strand DNA (55-MER), 12RSS signal top strand DNA (34-MER), 23RSS integration strand DNA (66-MER), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
3LC8
 
 | |
4F83
 
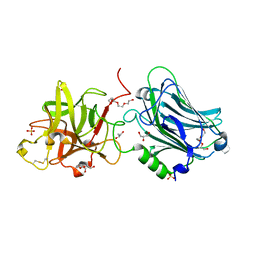 | | Crystal structure of the receptor binding domain of botulinum neurotoxin mosaic serotype C/D with a tetraethylene glycol molecule bound on the Hcn sub-domain and a sulfate ion at the putative active site | | Descriptor: | GLYCEROL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Zhang, Y, Buchko, G.W, Gardberg, A, Edwards, T.E, Sankaran, B, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-20 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the functional role of the Hcn sub-domain of the receptor-binding domain of the botulinum neurotoxin mosaic serotype C/D.
Biochimie, 95, 2013
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
1N6G
 
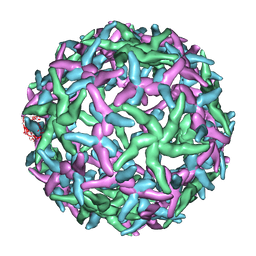 | | The structure of immature Dengue-2 prM particles | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
1NA4
 
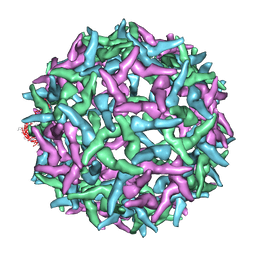 | | The structure of immature Yellow Fever virus particle | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
3OMW
 
 | | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II | | Descriptor: | CG14216 | | Authors: | Zhang, Y, Zhang, M, Zhang, Y. | | Deposit date: | 2010-08-27 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8701 Å) | | Cite: | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
3OMX
 
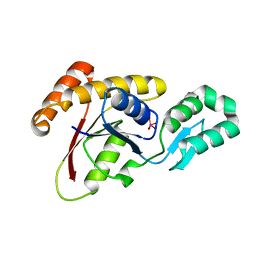 | | Crystal structure of Ssu72 with vanadate complex | | Descriptor: | CG14216, VANADATE ION | | Authors: | Zhang, Y, Zhang, M, Zhang, Y. | | Deposit date: | 2010-08-27 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3366 Å) | | Cite: | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
6XNZ
 
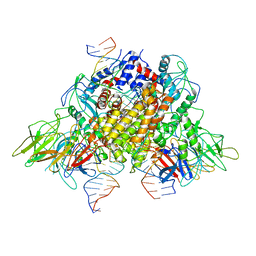 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Target Capture Complex | | Descriptor: | 12RSS integration strand (34-mer), 12RSS non-integration strand (34-mer), 23RSS integration strand (45-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
6Y5B
 
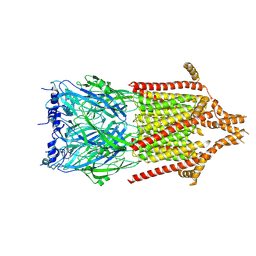 | | 5-HT3A receptor in Salipro (apo, asymmetric) | | Descriptor: | 5-hydroxytryptamine receptor 3A | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
6Y59
 
 | | 5-HT3A receptor in Salipro (apo, C5 symmetric) | | Descriptor: | 5-hydroxytryptamine receptor 3A | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
6Y5A
 
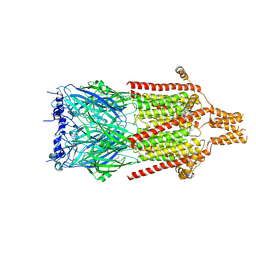 | | Serotonin-bound 5-HT3A receptor in Salipro | | Descriptor: | 5-hydroxytryptamine receptor 3A, SEROTONIN | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
6XNY
 
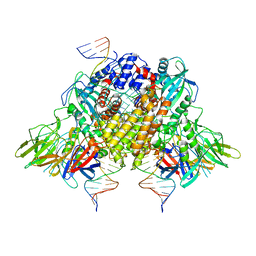 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Paired-Form) | | Descriptor: | 12RSS integration strand (55-mer), 12RSS signal DNA top strand (34-mer), 23RSS integration strand (66-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
1THD
 
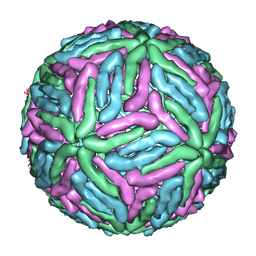 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | Descriptor: | Major envelope protein E | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
6O3X
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM2 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
6O3W
 
 | |
1O7Q
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | Deposit date: | 2002-11-12 | | Release date: | 2003-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
