1BGS
 
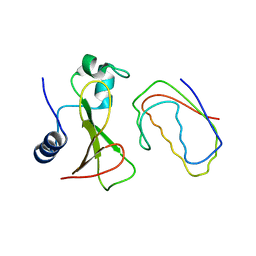 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | Descriptor: | BARNASE, BARSTAR | | Authors: | Guillet, V, Lapthorn, A, Mauguen, Y. | | Deposit date: | 1993-11-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
1CKE
 
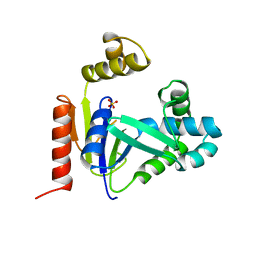 | | CMP KINASE FROM ESCHERICHIA COLI FREE ENZYME STRUCTURE | | Descriptor: | PROTEIN (CYTIDINE MONOPHOSPHATE KINASE), SULFATE ION | | Authors: | Briozzo, P, Golinelli-Pimpaneau, B. | | Deposit date: | 1998-09-24 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Structure, 6, 1998
|
|
3FKB
 
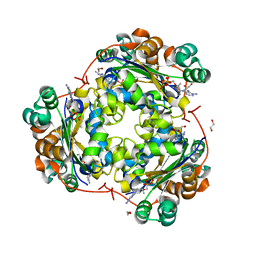 | | Structure of NDPK H122G and tenofovir-diphosphate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Morera, S, Chen, Y.X. | | Deposit date: | 2008-12-16 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleoside diphosphate kinase and the activation of antiviral phosphonate analogs of nucleotides: binding mode and phosphorylation of tenofovir derivatives
Nucleosides Nucleotides Nucleic Acids, 28, 2009
|
|
2AGK
 
 | |
2FA4
 
 | |
4ANE
 
 | | R80N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | CITRIC ACID, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Mycobacterium Tuberculosis Nucleoside Diphosphate Kinase R80N Mutant in Complex with Citrate
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ANC
 
 | | CRYSTAL FORM I OF THE D93N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intersubunit Ionic Interactions Stabilize the Nucleoside Diphosphate Kinase of Mycobacterium Tuberculosis.
Plos One, 8, 2013
|
|
4AND
 
 | | CRYSTAL FORM II OF THE D93N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Intersubunit Ionic Interactions Stabilize the Nucleoside Diphosphate Kinase of Mycobacterium Tuberculosis.
Plos One, 8, 2013
|
|
1BA7
 
 | | SOYBEAN TRYPSIN INHIBITOR | | Descriptor: | TRYPSIN INHIBITOR (KUNITZ) | | Authors: | De Meester, P, Brick, P, Lloyd, L.F, Blow, D.M, Onesti, S. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Kunitz-type soybean trypsin inhibitor (STI): implication for the interactions between members of the STI family and tissue-plasminogen activator.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2HUR
 
 | | Escherichia coli nucleoside diphosphate kinase | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Moynie, L, Giraud, M.-F, Georgescauld, F, Lascu, I, Dautant, A. | | Deposit date: | 2006-07-27 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of the Escherichia coli nucleoside diphosphate kinase reveals a new quaternary architecture for this enzyme family
Proteins, 67, 2007
|
|
3BBF
 
 | | Crystal structure of the NM23-H2 transcription factor complex with GDP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III1.
Mol.Cancer Ther., 8, 2009
|
|
3BBB
 
 | |
3BBC
 
 | |
2HVD
 
 | | Human nucleoside diphosphate kinase A complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Giraud, M.-F, Georgescauld, F, Lascu, I, Dautant, A. | | Deposit date: | 2006-07-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of S120G Mutant and Wild Type of Human Nucleoside Diphosphate Kinase A in Complex with ADP
J.Bioenerg.Biomembr., 38, 2006
|
|
2HVE
 
 | | S120G mutant of human nucleoside diphosphate kinase A complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Giraud, M.-F, Georgescauld, F, Lascu, I, Dautant, A. | | Deposit date: | 2006-07-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal Structures of S120G Mutant and Wild Type of Human Nucleoside Diphosphate Kinase A in Complex with ADP
J.Bioenerg.Biomembr., 38, 2006
|
|
1ODF
 
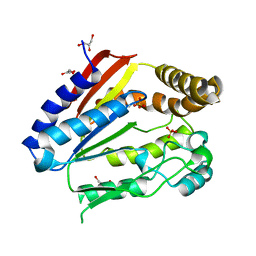 | | Structure of YGR205w protein. | | Descriptor: | GLYCEROL, HYPOTHETICAL 33.3 KDA PROTEIN IN ADE3-SER2 INTERGENIC REGION, SULFATE ION | | Authors: | Li De La Sierra-Gallay, I, Van Tilbeurgh, H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ygr205W Protein from Saccharomyces Cerevisiae: Close Structural Resemblance to E.Coli Pantothenate Kinase
Proteins: Struct.,Funct., Genet., 54, 2004
|
|
1QVZ
 
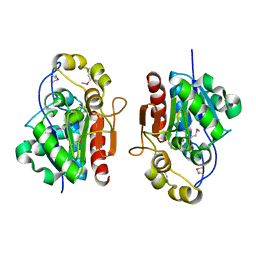 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QVV
 
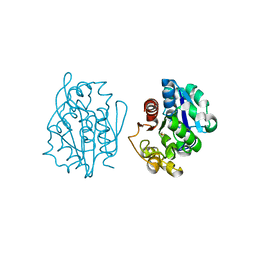 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1QVW
 
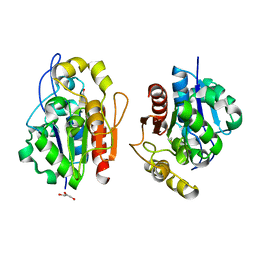 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | GLYCEROL, YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1K0D
 
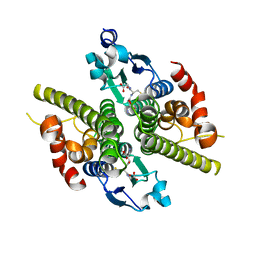 | | Ure2p in Complex with Glutathione | | Descriptor: | GLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1JZR
 
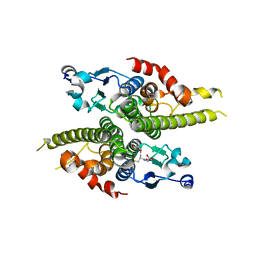 | | Ure2p in complex with glutathione | | Descriptor: | GLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-17 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1K0B
 
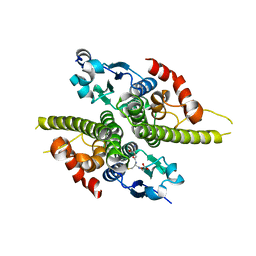 | | Ure2p in Complex with Glutathione | | Descriptor: | GLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1K0C
 
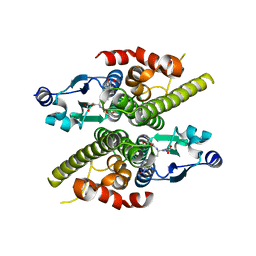 | | Ure2p in complex with S-p-nitrobenzylglutathione | | Descriptor: | GLUTATHIONE, S-(P-NITROBENZYL)GLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
1K0A
 
 | | Ure2p in Complex with S-hexylglutathione | | Descriptor: | GLUTATHIONE, S-HEXYLGLUTATHIONE, URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Melki, R, Morera, S. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds.
Biochemistry, 40, 2001
|
|
