2KKH
 
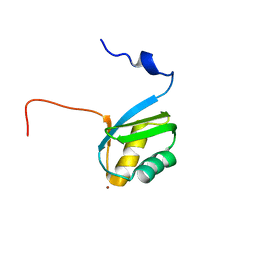 | | Structure of the zinc binding domain of the ATPase HMA4 | | Descriptor: | Putative heavy metal transporter, ZINC ION | | Authors: | Zimmerman, M, Clarke, O, Gulbis, J.M, Keizer, D.W, Jarvis, R.S, Cobbett, C.S, Hinds, M.G, Xiao, Z, Wedd, A.G. | | Deposit date: | 2009-06-20 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains
Biochemistry, 48, 2009
|
|
3DXS
 
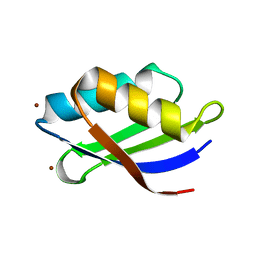 | | Crystal structure of a copper binding domain from HMA7, a P-type ATPase | | Descriptor: | Copper-transporting ATPase RAN1, LITHIUM ION, ZINC ION | | Authors: | Zimmermann, M, Xiao, Z, Clarke, O.B, Gulbis, J.M, Wedd, A.G. | | Deposit date: | 2008-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains.
Biochemistry, 48, 2009
|
|
2M37
 
 | | Structure of lasso peptide astexin-1 | | Descriptor: | ASTEXIN-1 | | Authors: | Zimmermann, M, Hegemann, J.D, Xie, X, Marahiel, M.A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The astexin-1 lasso peptides: biosynthesis, stability, and structural studies.
Chem.Biol., 20, 2013
|
|
2MLJ
 
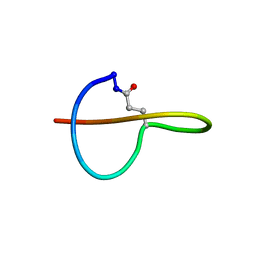 | |
9F8R
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (4-Chlorophenyl)[2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl]methanone | | Descriptor: | (4-chlorophenyl)-[2-(4-hydroxyphenyl)-1-benzothiophen-3-yl]methanone, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Zimmermann, M, Thiemann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
9F8P
 
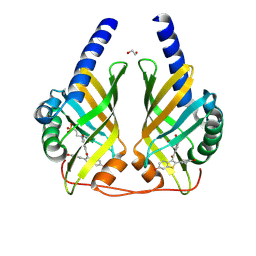 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with [6-Hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl](4-hydroxyphenyl)methanone | | Descriptor: | (4-hydroxyphenyl)-[2-(4-hydroxyphenyl)-6-oxidanyl-1-benzothiophen-3-yl]methanone, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Zimmermann, M, Thiemann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
9F8I
 
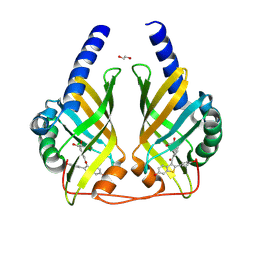 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (4-Chlorophenyl)[6-hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl]methanone | | Descriptor: | (4-chlorophenyl)-[2-(4-hydroxyphenyl)-6-oxidanyl-1-benzothiophen-3-yl]methanone, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Zimmermann, M, Thiemann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
9F8M
 
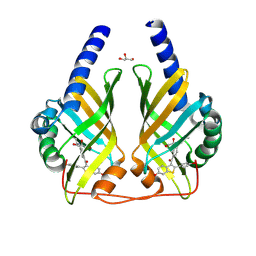 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with [6-Hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl][4-(propylamino)phenyl]methanone | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Phenazine biosynthesis protein A/B, ... | | Authors: | Zimmermann, M, Thiemann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
9F8S
 
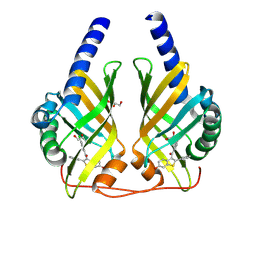 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (4-Chlorophenyl)[6-hydroxy-2-phenylbenzo[b]thiophen-3-yl]methanone | | Descriptor: | (4-chlorophenyl)-(6-oxidanyl-2-phenyl-1-benzothiophen-3-yl)methanone, GLYCEROL, Phenazine biosynthesis protein A/B | | Authors: | Zimmermann, M, Thiemann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
4JWV
 
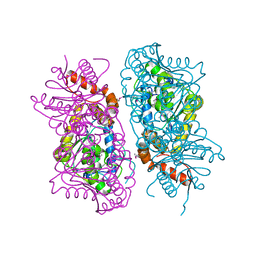 | | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Short chain enoyl-CoA hydratase | | Authors: | Cooper, D.R, Mikolajczak, K, Cymborowski, M, Grabowski, M, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-27 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4JVT
 
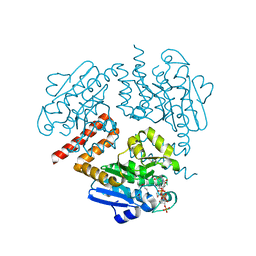 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase fromThermobifida fusca YX in complex with CoA | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, Enoyl-CoA hydratase | | Authors: | Mikolajczak, K, Porebski, P.J, Cooper, D.R, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase fromThermobifida fusca YX in complex with CoA
To be Published
|
|
4JSB
 
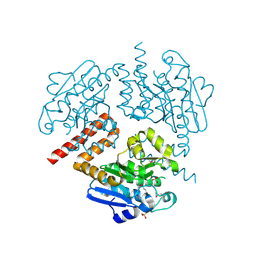 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX | | Descriptor: | Enoyl-CoA hydratase, SULFATE ION | | Authors: | Mikolajczak, K, Porebski, P.J, Cooper, D.R, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX
TO BE PUBLISHED
|
|
4JYJ
 
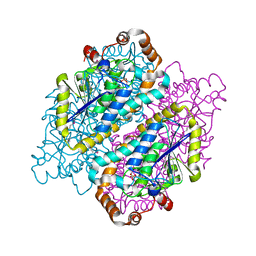 | | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Enoyl-CoA hydratase/isomerase, UNKNOWN LIGAND | | Authors: | Cooper, D.R, Porebski, P.J, Domagalski, M.J, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
3H5Q
 
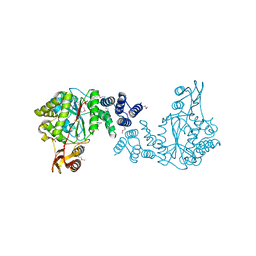 | | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus | | Descriptor: | Pyrimidine-nucleoside phosphorylase, SULFATE ION, THYMIDINE | | Authors: | Shumilin, I.A, Zimmerman, M, Cymborowski, M, Skarina, T, Onopriyenko, O, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus
TO BE PUBLISHED
|
|
8CQU
 
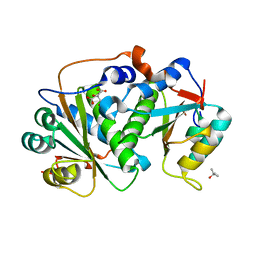 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1680) | | Descriptor: | CITRIC ACID, Putative NADH dehydrogenase/NAD(P)H nitroreductase, TERTIARY-BUTYL ALCOHOL | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQV
 
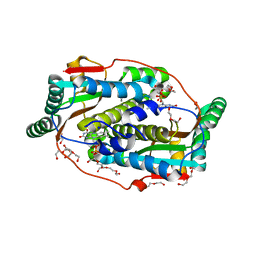 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_3392) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQT
 
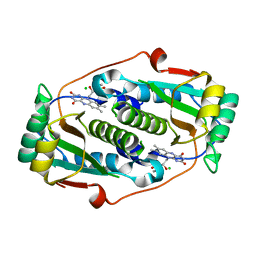 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_1316) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Putative NADH dehydrogenase/NAD(P)H nitroreductase | | Authors: | Blaha, J, Adam, L, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
8CQS
 
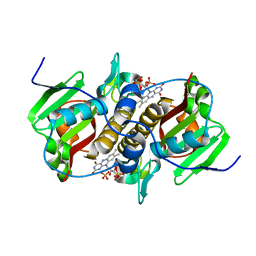 | | Flavin mononucleotide-dependent nitroreductase B.thetaiotaomicron (BT_0217) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase-like protein, PHOSPHATE ION | | Authors: | Blaha, J, Gratzl, S, Mortensen, S.A, Beckham, K.S.H, Chojnowski, G, Wilmanns, M, Zimmermann, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the diversity of nitroreductase enzymes in Bacteroides thetaiotaomicron
To Be Published
|
|
6EQC
 
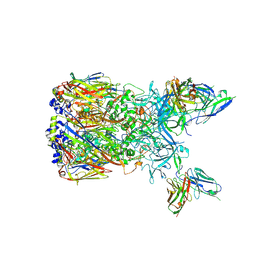 | | Cryo-EM reconstruction of a complex of a binding protein and human adenovirus C5 hexon | | Descriptor: | Hexon protein, scFv of 9C12 antibody | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
9O63
 
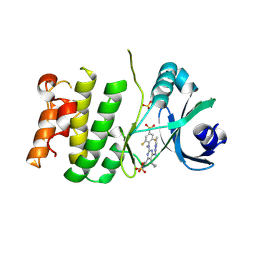 | | Crystal structure of PLK4 and RP1664 complex | | Descriptor: | 5-cyclopropyl-N~2~-[2,6-difluoro-4-(methanesulfonyl)phenyl]-N~2~-methyl-6-(1-methyl-1H-imidazol-4-yl)-N~4~-(5-methyl-1H-pyrazol-3-yl)pyrimidine-2,4-diamine, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Maderova, Z, Zimmermann, M, Sicheri, F. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of RP-1664: A First-in-Class Orally Bioavailable, Selective PLK4 Inhibitor.
J.Med.Chem., 2025
|
|
5OGI
 
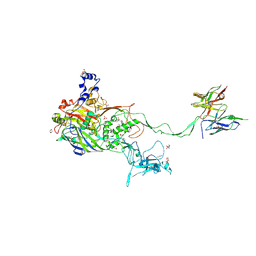 | | Complex of a binding protein and human adenovirus C 5 hexon | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
9F8N
 
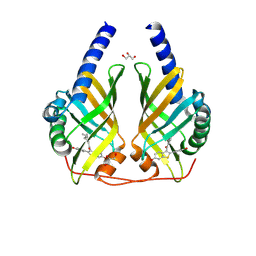 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with [6-Hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl][2-(isobutylamino)phenyl]methanone | | Descriptor: | GLYCEROL, Phenazine biosynthesis protein A/B, [2-(4-hydroxyphenyl)-6-oxidanyl-1-benzothiophen-3-yl]-[2-(2-methylpropylamino)phenyl]methanone | | Authors: | Thiemann, M, Zimmermann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
9F8Q
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with [6-Hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl](2-hydroxyphenyl)methanone | | Descriptor: | (2-hydroxyphenyl)-[2-(4-hydroxyphenyl)-6-oxidanyl-1-benzothiophen-3-yl]methanone, GLYCEROL, Phenazine biosynthesis protein A/B | | Authors: | Thiemann, M, Zimmermann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
9F8L
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with [6-Hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl](3-hydroxyphenyl)methanone | | Descriptor: | (3-hydroxyphenyl)-[2-(4-hydroxyphenyl)-6-oxidanyl-1-benzothiophen-3-yl]methanone, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Thiemann, M, Zimmermann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
9F8K
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with [6-Hydroxy-2-(4-hydroxyphenyl)benzo[b]thiophen-3-yl](3-isopropylphenyl)methanone | | Descriptor: | ACETATE ION, GLYCEROL, Phenazine biosynthesis protein A/B, ... | | Authors: | Thiemann, M, Zimmermann, M, Kunick, C, Blankenfeldt, W. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | From Bones to Bugs: Structure-Based Development of Raloxifene-Derived Pathoblockers That Inhibit Pyocyanin Production in Pseudomonas aeruginosa.
J.Med.Chem., 68, 2025
|
|
