7OBR
 
 | | RNC-SRP early complex | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Ban, N. | | Deposit date: | 2021-04-23 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
2A4R
 
 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4Q
 
 | | HCV NS3 protease with NS4a peptide and a covalently bound macrocyclic ketoamide compound. | | Descriptor: | (2R)-({N-[(3S)-3-({[(3S,6S)-6-CYCLOHEXYL-5,8-DIOXO-4,7-DIAZABICYCLO[14.3.1]ICOSA-1(20),16,18-TRIEN-3-YL]CARBONYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, NS3 protease/helicase', ... | | Authors: | Chen, K.X, Njoroge, F.G, Prongay, A, Pichardo, J, Madison, V, Girijavallabhan, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8TL6
 
 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF5 | | Descriptor: | DDB1- and CUL4-associated factor 5, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1 | | Authors: | Yue, H, Hunkeler, M, Roy Burman, S.S, Fischer, E.S. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Targeting DCAF5 suppresses SMARCB1-mutant cancer by stabilizing SWI/SNF.
Nature, 628, 2024
|
|
4DJV
 
 | | Structure of BACE Bound to 2-imino-5-(3'-methoxy-[1,1'-biphenyl]-3-yl)-3-methyl-5-phenylimidazolidin-4-one | | Descriptor: | (2E,5R)-2-imino-5-(3'-methoxybiphenyl-3-yl)-3-methyl-5-phenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DJY
 
 | |
4DJW
 
 | |
4DJX
 
 | |
4DJU
 
 | | Structure of BACE Bound to 2-imino-3-methyl-5,5-diphenylimidazolidin-4-one | | Descriptor: | (2E)-2-imino-3-methyl-5,5-diphenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3CQ0
 
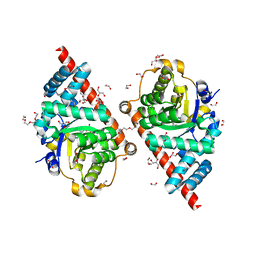 | | Crystal Structure of TAL2_YEAST | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative transaldolase YGR043C, ... | | Authors: | Huang, H, Niu, L, Teng, M. | | Deposit date: | 2008-04-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure and identification of NQM1/YGR043C, a transaldolase from Saccharomyces cerevisiae
Proteins, 73, 2008
|
|
3E17
 
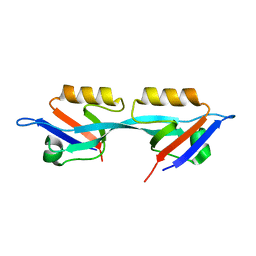 | | Crystal structure of the second PDZ domain from human Zona Occludens-2 | | Descriptor: | Tight junction protein ZO-2 | | Authors: | Chen, H, Tong, S.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-08-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the second PDZ domain from human zonula occludens 2
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1QUA
 
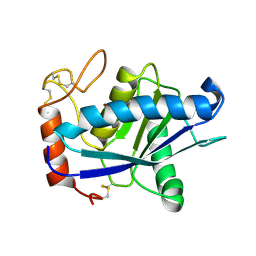 | | CRYSTAL STRUCTURE OF ACUTOLYSIN-C, A HEMORRHAGIC TOXIN FROM THE SNAKE VENOM OF AGKISTRODON ACUTUS, AT 2.2 A RESOLUTION | | Descriptor: | ACUTOLYSIN-C, ZINC ION | | Authors: | Niu, L, Teng, M, Zhu, X. | | Deposit date: | 1999-06-30 | | Release date: | 2000-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of acutolysin-C, a haemorrhagic toxin from the venom of Agkistrodon acutus, providing further evidence for the mechanism of the pH-dependent proteolytic reaction of zinc metalloproteinases.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6V9C
 
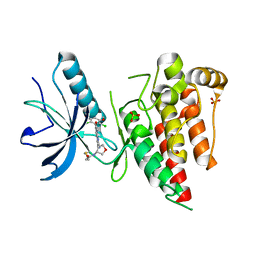 | | Crystal structure of FGFR4 kinase domain in complex with covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3R,4S)-4-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl]amino}oxolan-3-yl]prop-2-enamide, SULFATE ION | | Authors: | Liu, J, Liu, H. | | Deposit date: | 2019-12-13 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Selective, Covalent FGFR4 Inhibitors with Antitumor Activity in Models of Hepatocellular Carcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
5BT1
 
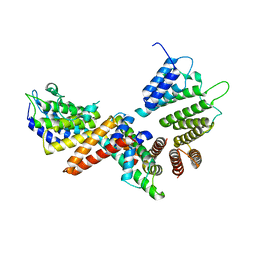 | | histone chaperone Hif1 playing with histone H2A-H2B dimer | | Descriptor: | HAT1-interacting factor 1, Histone H2A.1, Histone H2B.1 | | Authors: | Liu, H, Zhang, M, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2015-06-02 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Structure, 24, 2016
|
|
4R91
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(cyclopentylamino)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(1S,3R)-3-(cyclopentylamino)cyclohexyl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R92
 
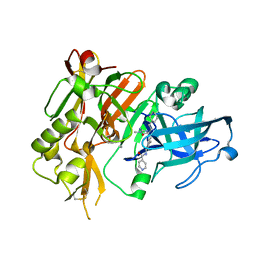 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(isonicotinamido)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]pyridine-4-carboxamide | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R95
 
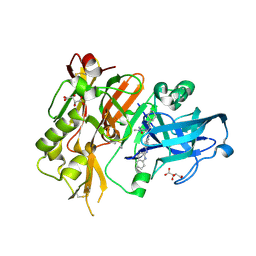 | | BACE-1 in complex with 2-(((1R,3S)-3-(((R)-4-(2-cyclohexylethyl)-2-iminio-1-methyl-5-oxoimidazolidin-4-yl)methyl)cyclohexyl)amino)quinolin-1-ium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-2-imino-3-methyl-5-{[(1S,3R)-3-(quinolin-2-ylamino)cyclohexyl]methyl}imidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R8Y
 
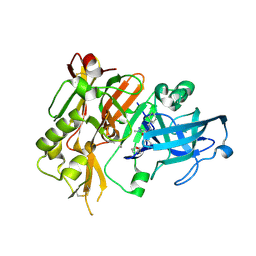 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((R)-1-(2-cyclopentylacetyl)pyrrolidin-3-yl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(3R)-1-(cyclopentylacetyl)pyrrolidin-3-yl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7NPL
 
 | | ALPHA-1 ANTITRYPSIN (C232S) COMPLEXED WITH cmpd 11 | | Descriptor: | Alpha-1-antitrypsin, GLYCEROL, N-((1S,2R)-1-(3-chloro-2-methylphenyl)-1-hydroxypentan-2-yl)-2-oxoindoline-4-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The development of highly potent and selective small molecule correctors of Z alpha 1 -antitrypsin misfolding.
Bioorg.Med.Chem.Lett., 41, 2021
|
|
7NPK
 
 | | ALPHA-1 ANTITRYPSIN C232S COMPLEXED WITH CMPD3 | | Descriptor: | Alpha-1-antitrypsin, GLYCEROL, N-((1S,2R)-1-hydroxy-1-(o-tolyl)pentan-2-yl)-2-oxo-2,3-dihydrobenzo[d]oxazole-5-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The development of highly potent and selective small molecule correctors of Z alpha 1 -antitrypsin misfolding.
Bioorg.Med.Chem.Lett., 41, 2021
|
|
4R93
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-1-methyl-5-oxo-4-(((1S,3R)-3-(3-phenylureido)cyclohexyl)methyl)imidazolidin-2-iminium | | Descriptor: | 1-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]-3-phenylurea, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7NFQ
 
 | | Fujian capmidlink domain in complex with Nb8193 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nb8193, ... | | Authors: | Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIL
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8190 core | | Descriptor: | Nanobody8190 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-12 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIS
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8192 core | | Descriptor: | Nanobody8192 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIR
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8191 core | | Descriptor: | Nanobody8191 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
