6M5V
 
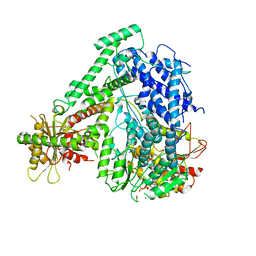 | | The coordinate of the hexameric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
7UOM
 
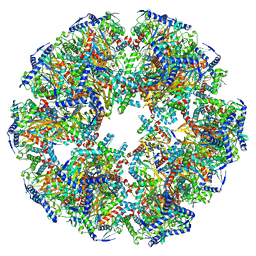 | | Endogenous dihydrolipoamide acetyltransferase (E2) core of pyruvate dehydrogenase complex from bovine kidney | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Liu, S, Xia, X, Zhen, J, Li, Z.H, Zhou, Z.H. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney.
Cell Discov, 8, 2022
|
|
7UOL
 
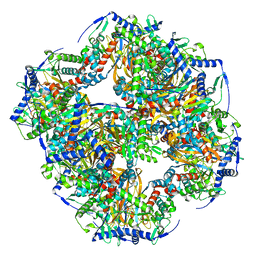 | | Endogenous dihydrolipoamide succinyltransferase (E2) core of 2-oxoglutarate dehydrogenase complex from bovine kidney | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Liu, S, Xia, X, Zhen, J, Li, Z.H, Zhou, Z.H. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney.
Cell Discov, 8, 2022
|
|
6NSJ
 
 | | CryoEM structure of Helicobacter pylori urea channel in closed state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | Authors: | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6NSK
 
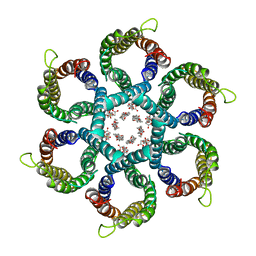 | | CryoEM structure of Helicobacter pylori urea channel in open state. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | Authors: | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
8FJL
 
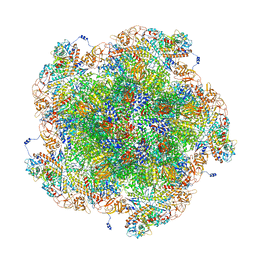 | | Golden Shiner Reovirus Core Tropical Vertex | | Descriptor: | Clamp protein VP6, Major inner capsid protein VP3, Microtubule-associated protein VP5, ... | | Authors: | Stevens, A.S, Zhou, Z.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Asymmetric reconstruction of the aquareovirus core at near-atomic resolution and mechanism of transcription initiation.
Protein Cell, 14, 2023
|
|
8FJP
 
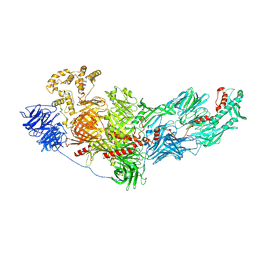 | | Cryo-EM structure of native mosquito salivary gland surface protein 1 (SGS1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, salivary gland surface protein 1 | | Authors: | Liu, S, Xia, X, Calvo, E, Zhou, Z.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Native structure of mosquito salivary protein uncovers domains relevant to pathogen transmission.
Nat Commun, 14, 2023
|
|
8FJK
 
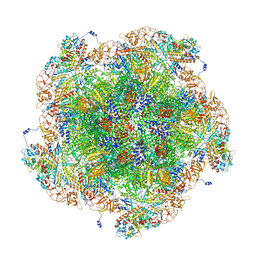 | | Golden Shiner Reovirus Core Polar Vertex | | Descriptor: | Clamp protein VP6, Major inner capsid protein VP3, Microtubule-associated protein VP5, ... | | Authors: | Stevens, A.S, Zhou, Z.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-22 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Asymmetric reconstruction of the aquareovirus core at near-atomic resolution and mechanism of transcription initiation.
Protein Cell, 14, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
7UWS
 
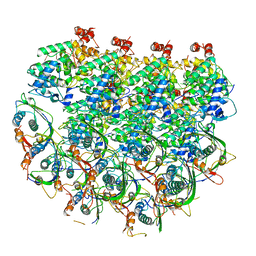 | | Atomic model of the partial VSV nucleocapsid | | Descriptor: | Matrix protein, Nucleoprotein, RNA | | Authors: | Zhou, K, Si, Z, Ge, P, Tsao, J, Luo, M, Zhou, Z.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Atomic model of vesicular stomatitis virus and mechanism of assembly.
Nat Commun, 13, 2022
|
|
6M5U
 
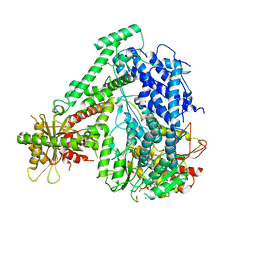 | | The coordinates of the monomeric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Zhu, L, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
7TW2
 
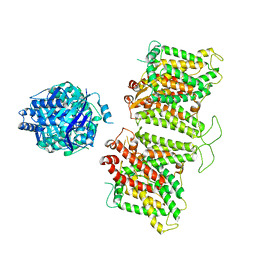 | |
7TW1
 
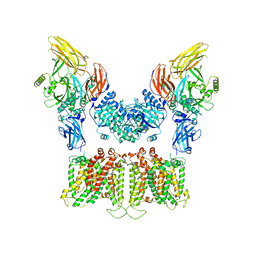 | |
7TVZ
 
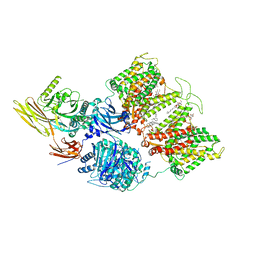 | | Cryo-EM structure of human band 3-protein 4.2 complex in diagonal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW0
 
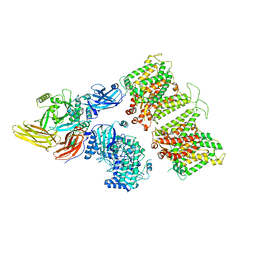 | | Cryo-EM structure of human band 3-protein 4.2 complex in vertical conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW5
 
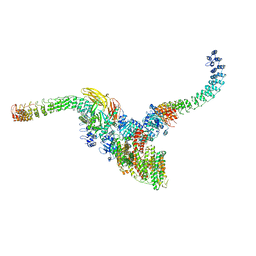 | | Cryo-EM structure of human ankyrin complex (B2P1A2) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW6
 
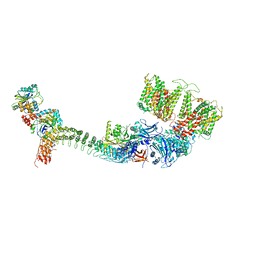 | | Cryo-EM structure of human ankyrin complex (B4P1A1) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW3
 
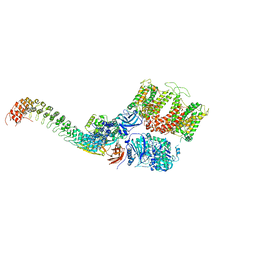 | | Cryo-EM structure of human ankyrin complex (B2P1A1) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8FVI
 
 | | Human APOBEC3H bound to HIV-1 Vif in complex with CBF-beta, ELOB, ELOC, and CUL5 | | Descriptor: | Core-binding factor subunit beta, Cullin 5, DNA dC->dU-editing enzyme APOBEC-3H, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Nat Commun, 14, 2023
|
|
8FVJ
 
 | | Dimeric form of HIV-1 Vif in complex with human CBF-beta, ELOB, ELOC, and CUL5 | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Elongin-B, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Nat Commun, 14, 2023
|
|
