2IHX
 
 | |
6KAB
 
 | |
6KCN
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 4 | | Descriptor: | 3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)isochromen-1-one, GLYCEROL, LYSINE, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-28 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
6KA6
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 1 | | Descriptor: | (3~{S})-3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, GLYCEROL, LYSINE, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-21 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
6KCT
 
 | |
6KBF
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 3 | | Descriptor: | (3~{S})-3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)-3,4-dihydro-2~{H}-isoquinolin-1-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
3LWV
 
 | | Structure of H/ACA RNP bound to a substrate RNA containing 2'-deoxyuridine | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(du)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
3LWO
 
 | | Structure of H/ACA RNP bound to a substrate RNA containing 5BrU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(5BU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study.
J.Mol.Biol., 401, 2010
|
|
3LWP
 
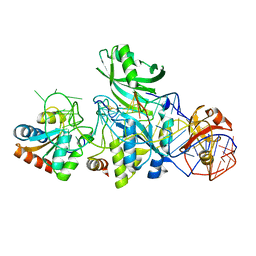 | | Structure of H/ACA RNP bound to a substrate RNA containing 5BrdU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(BRU)P*GP*CP*GP*GP*UP*UP*U)-3, 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study.
J.Mol.Biol., 401, 2010
|
|
3LWR
 
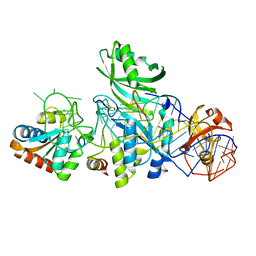 | | Structure of H/ACA RNP bound to a substrate RNA containing 4SU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(4SU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
3LWQ
 
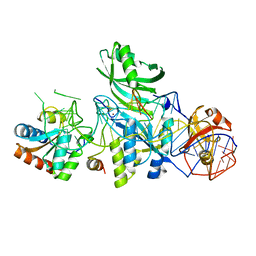 | | Structure of H/ACA RNP bound to a substrate RNA containing 3MU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(UR3)P*GP*CP*GP*GP*UP*UP*U)-3, 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.678 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
3MQK
 
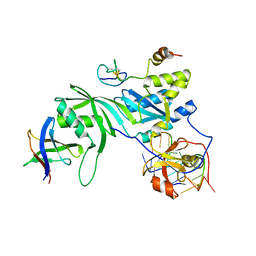 | | Cbf5-Nop10-Gar1 complex binding with 17mer RNA containing ACA trinucleotide | | Descriptor: | RNA (5'-R(P*CP*GP*AP*UP*CP*CP*AP*CP*A)-3'), RNA (5'-R(P*GP*UP*UP*CP*GP*AP*UP*CP*CP*AP*CP*AP*G)-3'), Ribosome biogenesis protein Nop10, ... | | Authors: | Zhou, J, Liang, B, Li, H. | | Deposit date: | 2010-04-28 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional evidence of high specificity of Cbf5 for ACA trinucleotide.
Rna, 17, 2011
|
|
7WUP
 
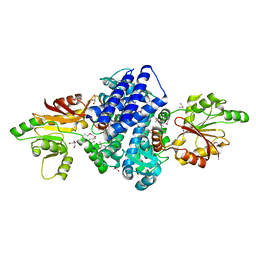 | | The crystal structure of ApiI | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ApiI, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-09 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of ApiI
To Be Published
|
|
7WW0
 
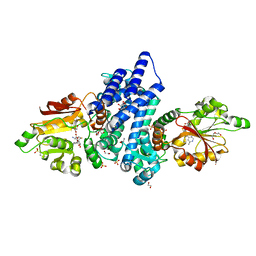 | | The crystal structure of FinI complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of FinI complex with SAH
To Be Published
|
|
8HS0
 
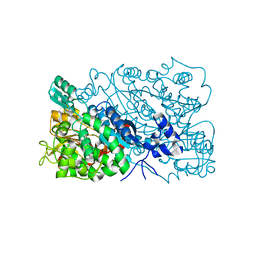 | | The mutant structure of DHAD V178W | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The mutant structure of DHAD
To Be Published
|
|
8IMU
 
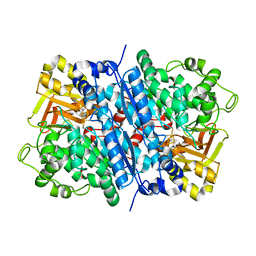 | | Dihydroxyacid dehydratase (DHAD) mutant-V497F | | Descriptor: | ACETATE ION, Dihydroxy-acid dehydratase, chloroplastic, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dihydroxyacid dehydratase (DHAD) mutant-V497F
To Be Published
|
|
8IKZ
 
 | | The mutant structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The mutant structure of DHAD
To Be Published
|
|
7WUY
 
 | | The crystal structure of FinI in complex with SAM and fischerin | | Descriptor: | 1,2-ETHANEDIOL, 3-[[(1~{S},2~{R},4~{a}~{S},8~{a}~{S})-2-methyl-1,2,4~{a},5,6,7,8,8~{a}-octahydronaphthalen-1-yl]carbonyl]-5-[(1~{S},2~{R},5~{S},6~{S})-2,5-bis(oxidanyl)-7-oxabicyclo[4.1.0]heptan-2-yl]-4-oxidanyl-1~{H}-pyridin-2-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The crystal structure of FinI in complex with SAM and fischerin
To Be Published
|
|
7X2S
 
 | | Crystal Structure of hetero-Diels-Alderase PycR1 in complex with Neosetophomone B and tropolone o-quinone methide | | Descriptor: | (1~{S},3~{R},11~{R},13~{S},14~{Z},18~{E})-1,8,14,17,17-pentamethyl-5,13-bis(oxidanyl)-2-oxatricyclo[9.9.0.0^{3,9}]icosa-4,7,9,14,18-pentaen-6-one, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Calcium-dependent glycosylated enzyme in the tandem hetero-Diels-Alder reaction
To Be Published
|
|
7X2X
 
 | | Crystal Structure of hetero-Diels-Alderase PycR1 in complex with 10-hydroxy-8E-humulene | | Descriptor: | (1R,2E,6E,9E)-2,5,5,9-tetramethylcycloundeca-2,6,9-trien-1-ol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Calcium-dependent glycosylated enzyme in the tandem hetero-Diels-Alder reaction
To Be Published
|
|
7X2N
 
 | | Crystal structure of Diels-Alderase PycR1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of hetero-Diels-Alderase PycR1
To Be Published
|
|
7X36
 
 | | Crystal Structure of hetero-Diels-Alderase EupfF | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-28 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Calcium-dependent glycosylated enzyme in the tandem hetero-Diels-Alder reaction
To Be Published
|
|
8GRX
 
 | | APOE4 receptor in complex with APOE4 NTD | | Descriptor: | Apolipoprotein E, Leukocyte immunoglobulin-like receptor subfamily A member 6 | | Authors: | Zhou, J, Wang, Y, Huang, G, Shi, Y. | | Deposit date: | 2022-09-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | LilrB3 is a putative cell surface receptor of APOE4.
Cell Res., 33, 2023
|
|
7E40
 
 | | Mechanism of Phosphate Sensing and Signaling Revealed by Rice SPX1-PHR2 Complex Structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 1,Endolysin | | Authors: | Zhou, J, Hu, Q, Yao, D, Xing, W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure.
Nat Commun, 12, 2021
|
|
7ETR
 
 | |
