7DU2
 
 | | RNA polymerase III EC complex in post-translocation state | | 分子名称: | DNA (5'-D(P*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*T)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | 著者 | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | 登録日 | 2021-01-07 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
7EGF
 
 | | TFIID lobe A subcomplex | | 分子名称: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 11, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 2021
|
|
7EGG
 
 | | TFIID lobe B subcomplex | | 分子名称: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 4, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGH
 
 | | TFIID lobe C subcomplex | | 分子名称: | DNA (45-MER), Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 2, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGI
 
 | | TFIID in rearranged conformation | | 分子名称: | TATA-box-binding protein, Transcription initiation factor IIA subunit 1, Transcription initiation factor IIA subunit 2, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (9.82 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGD
 
 | | SCP promoter-bound TFIID-TFIIA in initial TBP-loading state | | 分子名称: | DNA (72-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (6.75 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGJ
 
 | | SCP promoter-bound TFIID-TFIIA in post TBP-loading state | | 分子名称: | DNA (74-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (8.64 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGE
 
 | | TFIID in canonical conformation | | 分子名称: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | 著者 | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7F4G
 
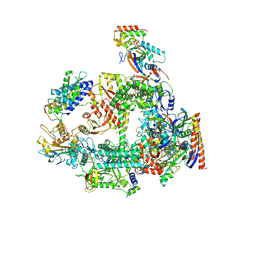 | | Structure of RPAP2-bound RNA polymerase II | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Chen, X, Qi, Y, Wang, X, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-06-18 | | 公開日 | 2021-07-07 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (2.78 Å) | | 主引用文献 | RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex.
Cell Rep, 39, 2022
|
|
6LTK
 
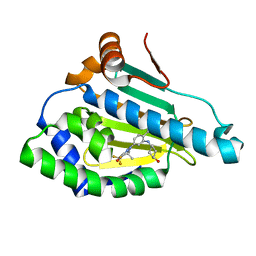 | | HSP90 in complex with SNX-2112 | | 分子名称: | 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein HSP 90-alpha | | 著者 | Cao, H.L. | | 登録日 | 2020-01-22 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.141 Å) | | 主引用文献 | Complex Crystal Structure Determination and in vitro Anti-non-small Cell Lung Cancer Activity of Hsp90 N Inhibitor SNX-2112.
Front Cell Dev Biol, 9, 2021
|
|
1CO0
 
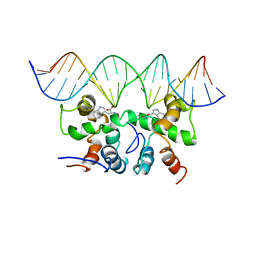 | | NMR STUDY OF TRP REPRESSOR-MTR OPERATOR DNA COMPLEX | | 分子名称: | 5'-D(*TP*GP*TP*AP*CP*CP*AP*GP*TP*AP*CP*AP*CP*GP*AP*GP*TP*AP*CP*A)-3', 5'-D(*TP*GP*TP*AP*CP*TP*CP*GP*TP*GP*TP*AP*CP*TP*GP*GP*TP*AP*CP*A)-3', TRP OPERON REPRESSOR, ... | | 著者 | Zhou, G.P, Brocchieri, L, Jardetzky, O. | | 登録日 | 1999-05-30 | | 公開日 | 2003-09-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Allostery and Induced Fit, NMR and Molecular Modeling Study of the
trp-repressor - mtr DNA complex
Structures and Mechanisms, ACS Symposium Series, 827, 2002
|
|
7BQA
 
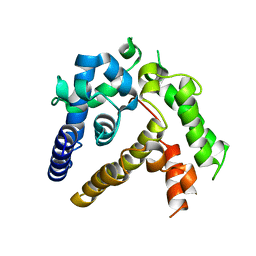 | | Crystal structure of ASFV p35 | | 分子名称: | 60 kDa polyprotein | | 著者 | Li, G.B, Fu, D, Chen, C, Guo, Y. | | 登録日 | 2020-03-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2021-05-05 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | 分子名称: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.847 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | 分子名称: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVF
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | 分子名称: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.655 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
8UQ9
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 4-residue linker | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ8
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 2-residue linker | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQA
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 12-residue linker | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, SODIUM ION, ... | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.049 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQC
 
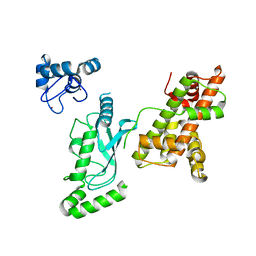 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 2) | | 分子名称: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQB
 
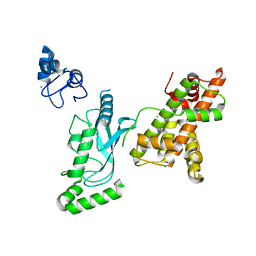 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 1) | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.484 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQD
 
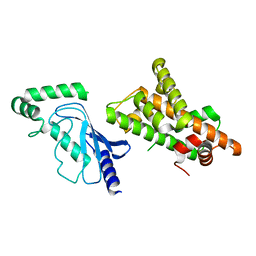 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (condition 2. RING not modeled in density) | | 分子名称: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.893 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQE
 
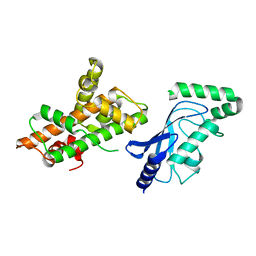 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 26-residue linker (RING not modeled in density) | | 分子名称: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.562 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
6CO2
 
 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | 分子名称: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | 著者 | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | 登録日 | 2018-03-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CO1
 
 | |
6D0L
 
 | | Structure of human TIRR | | 分子名称: | Tudor-interacting repair regulator protein | | 著者 | Cui, G, Botuyan, M.V, Mer, G. | | 登録日 | 2018-04-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
