3PY3
 
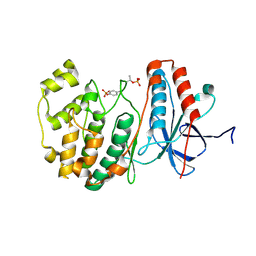 | |
6A9D
 
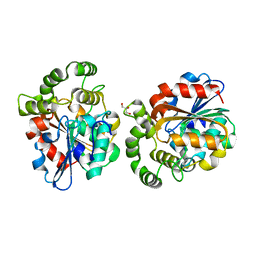 | |
3TG3
 
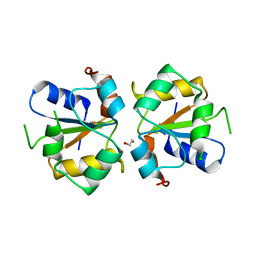 | | Crystal structure of the MAPK binding domain of MKP7 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 16 | | Authors: | Zhang, Y.Y, Liu, X, Wu, J.W, Wang, Z.X. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | A Distinct Interaction Mode Revealed by the Crystal Structure of the Kinase p38alpha with the MAPK Binding Domain of the Phosphatase MKP5.
Sci.Signal., 4, 2011
|
|
3TG1
 
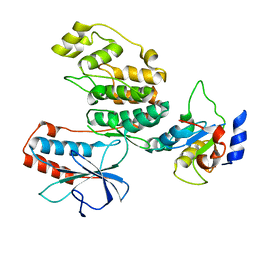 | |
8YPF
 
 | | The crystal structure of inactive p38 complexed with a ATF2 from 46 to 90 | | Descriptor: | Cyclic AMP-dependent transcription factor ATF-2, Mitogen-activated protein kinase 14 | | Authors: | Zhang, Y.Y, Pei, C.J, He, Q.Q, Luo, Z.P, Wu, J.W, Wang, Z.X. | | Deposit date: | 2024-03-16 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic insights into the allosteric activation of p38alpha MAP kinase via specific docking interactions
To Be Published
|
|
8YPE
 
 | | The crystal structure of inactive p38 complexed with a ATF2 from 46 to 80 | | Descriptor: | Cyclic AMP-dependent transcription factor ATF-2, Mitogen-activated protein kinase 14 | | Authors: | Zhang, Y.Y, Pei, C.J, He, Q.Q, Luo, Z.P, Wu, J.W, Wang, Z.X. | | Deposit date: | 2024-03-16 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic insights into the allosteric activation of p38alpha MAP kinase via specific docking interactions
To Be Published
|
|
8YP8
 
 | | Structure of the p38alpha-pepHePTPm(16-31)(V31C ) complex | | Descriptor: | Mitogen-activated protein kinase 14, Tyrosine-protein phosphatase non-receptor type 7 | | Authors: | Zhang, Y.Y, Pei, C.J, He, Q.Q, Luo, Z.P, Wu, J.W, Wang, Z.X. | | Deposit date: | 2024-03-15 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and mechanistic insights into the allosteric activation of p38alpha MAP kinase via specific docking interactions
To Be Published
|
|
3JAU
 
 | | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | | Descriptor: | Capsid protein VP1, Heavy chain of Fab fragment variable region of antibody D5, Light chain of Fab fragment variable region of antibody D5 | | Authors: | Fan, C, Ye, X.H, Ku, Z.Q, Zuo, T, Kong, L.L, Zhang, C, Shi, J.P, Liu, Q.W, Chen, T, Zhang, Y.Y, Jiang, W, Zhang, L.Q, Huang, Z, Cong, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Recognition of Human Enterovirus 71 by a Bivalent Broadly Neutralizing Monoclonal Antibody
Plos Pathog., 12, 2016
|
|
4F2L
 
 | | Structure of a regulatory domain of AMPK | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, MAGNESIUM ION | | Authors: | Xin, F.J, Zhang, Y.Y, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2012-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved regulatory elements in AMPK
Nature, 498, 2013
|
|
9JVX
 
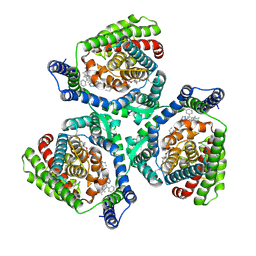 | | Overall structure of human EAAT2 bound with activator (GT949) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 3-[(~{R})-(4-cyclohexylpiperazin-1-yl)-[1-(2-phenylethyl)-1,2,3,4-tetrazol-5-yl]methyl]-6-methoxy-1~{H}-quinolin-2-one, CHOLESTEROL, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Shi, Y, Huang, J, Zhou, Q. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural study of human glutamate transporter EAAT2
Dian Zi Xian Wei Xue Bao, 2025
|
|
9JVW
 
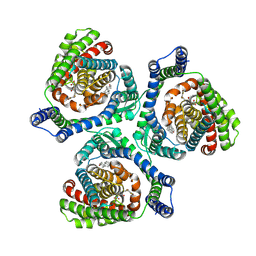 | | Overall structure of human EAAT2 bound with inhibitor (WAY-213613) | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, CHOLESTEROL, Excitatory amino acid transporter 2 | | Authors: | Xia, L.Y, Zhang, Y.Y, Shi, Y, Huang, J, Zhou, Q. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural study of human glutamate transporter EAAT2
Dian Zi Xian Wei Xue Bao, 2025
|
|
9JVV
 
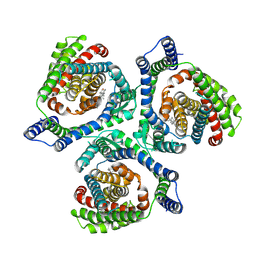 | | Overall structure of human EAAT2 in the substrate-free state | | Descriptor: | CHOLESTEROL, Excitatory amino acid transporter 2 | | Authors: | Xia, L.Y, Zhang, Y.Y, Shi, Y, Huang, J, Zhou, Q. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural study of human glutamate transporter EAAT2
Dian Zi Xian Wei Xue Bao, 2025
|
|
7XUJ
 
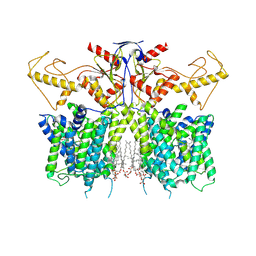 | | Human SLC26A3 in complex with UK5099 | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | human SLC26A3 in complex with UK5099
To Be Published
|
|
7XUL
 
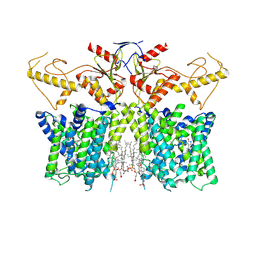 | | Human SLC26A3 in complex with Tenidap | | Descriptor: | 5-chloranyl-2-oxidanyl-3-thiophen-2-ylcarbonyl-indole-1-carboxamide, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of human SLC26A3 in complex with Tenidap
To Be Published
|
|
7XUH
 
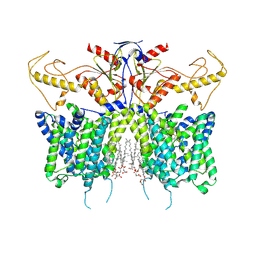 | | Down-regulated in adenoma in complex with TQR1122 | | Descriptor: | 2-[4,8-dimethyl-2-oxidanylidene-7-[[3-(trifluoromethyl)phenyl]methoxy]chromen-3-yl]ethanoic acid, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | structure of human SLC26A3 in complex with TQR1122
To Be Published
|
|
8JD7
 
 | |
8IET
 
 | |
8XL9
 
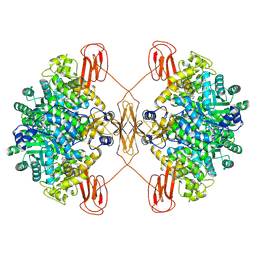 | | Structure of human pyruvate carboxylase | | Descriptor: | BIOTIN, Pyruvate carboxylase, mitochondrial | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC).
Elife, 2024
|
|
8HLD
 
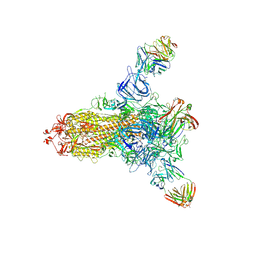 | | S protein of SARS-CoV-2 in complex with 26434 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
MedComm (2020), 5, 2024
|
|
8HLC
 
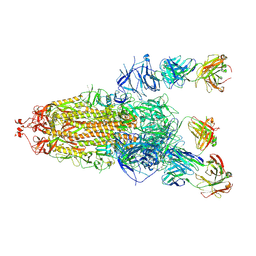 | | S protein of SARS-CoV-2 in complex with 3711 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
MedComm (2020), 5, 2024
|
|
7D0A
 
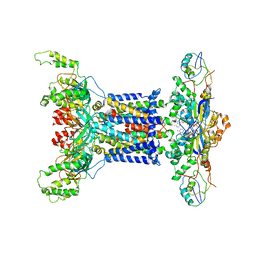 | | Acinetobacter MlaFEDB complex in ADP-vanadate trapped Vclose conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D09
 
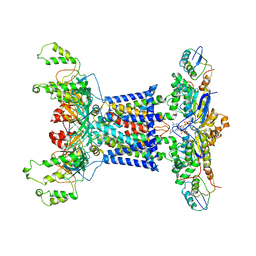 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans2 conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D08
 
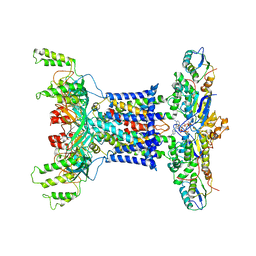 | | Acinetobacter MlaFEDB complex in ATP-bound Vtrans1 conformation | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
7D06
 
 | | Cryo EM structure of the nucleotide free Acinetobacter MlaFEDB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ABC transporter ATP-binding protein, Anti-sigma factor antagonist, ... | | Authors: | Zhang, Y.Y, Fan, Q.X, Chi, X.M, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-09-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.
Cell Discov, 6, 2020
|
|
6J2R
 
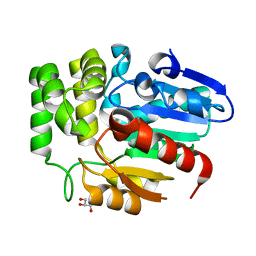 | | Crystal structure of Striga hermonthica HTL8 (ShHTL8) | | Descriptor: | GLYCEROL, Hyposensitive to light 8 | | Authors: | Zhang, Y.Y, Xi, Z. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and biochemical characterization of Striga hermonthica HYPO-SENSITIVE TO LIGHT 8 (ShHTL8) in strigolactone signaling pathway.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
