110L
 
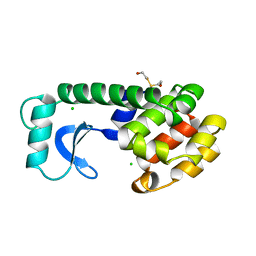 | |
111L
 
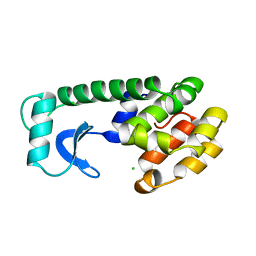 | |
113L
 
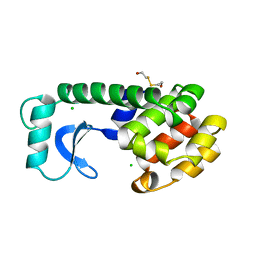 | |
115L
 
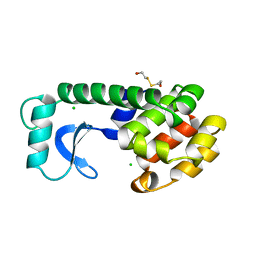 | |
114L
 
 | |
137L
 
 | |
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | 登録日 | 1991-01-25 | | 公開日 | 1992-07-15 | | 最終更新日 | 2021-06-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
258L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, ZINC ION | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-01-05 | | 公開日 | 2000-09-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
216L
 
 | |
260L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, NICKEL (II) ION, PROTEIN (LYSOZYME) | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-03-01 | | 公開日 | 2000-09-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
259L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, COBALT (II) ION, PROTEIN (LYSOZYME) | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-02-10 | | 公開日 | 1999-04-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
217L
 
 | |
257L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-01-05 | | 公開日 | 2000-09-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
7CY6
 
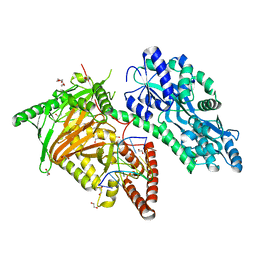 | | Crystal Structure of CMD1 in complex with 5mC-DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY7
 
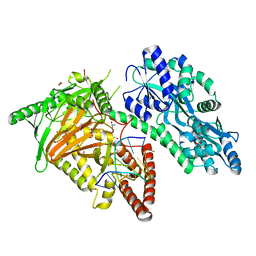 | | Crystal Structure of CMD1 in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY5
 
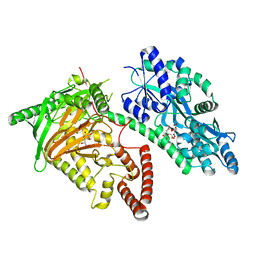 | | Crystal Structure of CMD1 in complex with vitamin C | | 分子名称: | ASCORBIC ACID, CITRIC ACID, FE (III) ION, ... | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY8
 
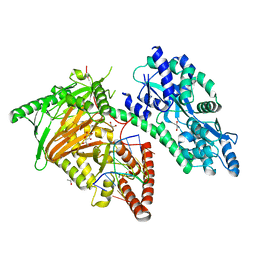 | | Crystal Structure of CMD1 in complex with 5mC-DNA and vitamin C | | 分子名称: | 1,2-ETHANEDIOL, ASCORBIC ACID, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), ... | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY4
 
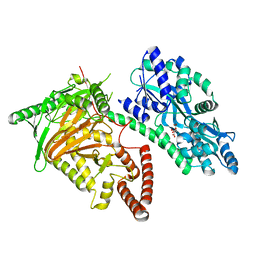 | | Crystal Structure of CMD1 in apo form | | 分子名称: | CITRIC ACID, FE (III) ION, Maltodextrin-binding protein,5-methylcytosine-modifying enzyme 1 | | 著者 | Li, W, Zhang, T, Sun, M, Ding, J. | | 登録日 | 2020-09-03 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
1YFJ
 
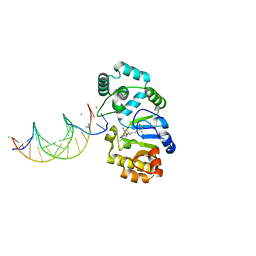 | | T4Dam in Complex with AdoHcy and 15-mer Oligonucleotide Showing Semi-specific and Specific Contact | | 分子名称: | 5'-D(*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*G)-3', CALCIUM ION, CHLORIDE ION, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-02 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YF3
 
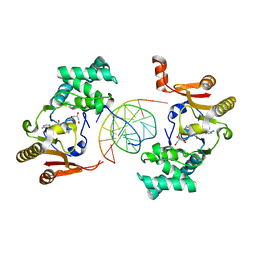 | | T4Dam in Complex with AdoHcy and 13-mer Oligonucleotide Making Non- and Semi-specific (~1/4) Contact | | 分子名称: | 5'-D(*AP*CP*CP*AP*TP*GP*AP*TP*CP*TP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*CP*AP*TP*GP*G)-3', DNA adenine methylase, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2004-12-30 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YFL
 
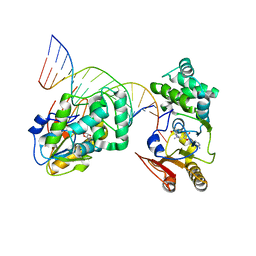 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | 分子名称: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-03 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
5XPD
 
 | |
235L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | 著者 | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | 登録日 | 1997-10-17 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
242L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | 著者 | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | 登録日 | 1997-10-23 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
240L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | 著者 | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | 登録日 | 1997-10-22 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
