4RLV
 
 | |
4RLY
 
 | |
2FQD
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQG
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQE
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQF
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
4D8O
 
 | |
2IDG
 
 | | Crystal Structure of hypothetical protein AF0160 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0160 | | Authors: | Zhao, M, Zhang, M, Chang, J, Chen, L, Xu, H, Li, Y, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Hypothetical Protein AF0160 from Archaeoglobus fulgidus at 2.69 Angstrom resolution
To be Published
|
|
4DC2
 
 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | Descriptor: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | Authors: | Shang, Y, Wang, C, Yu, J, Zhang, M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
4EGX
 
 | | Crystal structure of KIF1A CC1-FHA tandem | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Kinesin-like protein KIF1A | | Authors: | Yu, J, Huo, L, Yue, Y, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-02 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
4WSI
 
 | | Crystal Structure of PALS1/Crb complex | | Descriptor: | MAGUK p55 subfamily member 5, Protein crumbs | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1IOU
 
 | |
4YL8
 
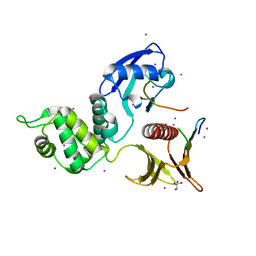 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
4YTL
 
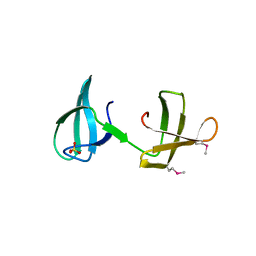 | | Structure of the KOW2-KOW3 Domain of Transcription Elongation Factor Spt5. | | Descriptor: | GLYCEROL, SULFATE ION, Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, ZHang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
4YTK
 
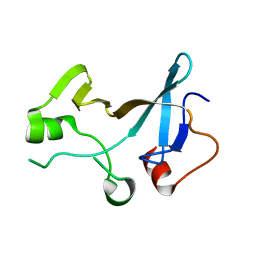 | | Structure of the KOW1-Linker1 domain of Transcription Elongation Factor Spt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, Zhang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.0904 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
4ZRK
 
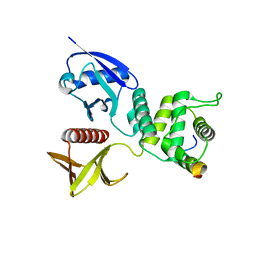 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
1PWJ
 
 | | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of Domain Swapped Dimer Assembly | | Descriptor: | dynein light chain-2 | | Authors: | Wang, W, Lo, K.W.-H, Kan, H.-M, Fan, J.-S, Zhang, M. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of the Domain-swapped Dimer Assembly
J.Biol.Chem., 278, 2003
|
|
1PWK
 
 | | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of Domain Swapped Dimer Assembly | | Descriptor: | dynein light chain-2 | | Authors: | Wang, W, Lo, K.W.-H, Kan, H.-M, Fan, J.-S, Zhang, M. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the Monomeric 8-kDa Dynein Light Chain and Mechanism of the Domain-swapped Dimer Assembly
J.Biol.Chem., 278, 2003
|
|
1Q5F
 
 | | NMR Structure of Type IVb pilin (PilS) from Salmonella typhi | | Descriptor: | PilS | | Authors: | Xu, X.F, Tan, Y.W, Hackett, J, Zhang, M, Mok, Y.K. | | Deposit date: | 2003-08-07 | | Release date: | 2004-07-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Type IVb Pilin from Salmonella typhi and Its Assembly into Pilus
J.Biol.Chem., 279, 2004
|
|
5WBX
 
 | |
5WC5
 
 | |
1RSO
 
 | | Hetero-tetrameric L27 (Lin-2, Lin-7) domain complexes as organization platforms of supra-molecular assemblies | | Descriptor: | Peripheral plasma membrane protein CASK, Presynaptic protein SAP97 | | Authors: | Feng, W, Long, J.-F, Fan, J.-S, Suetake, T, Zhang, M. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-04 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | The tetrameric L27 domain complex as an organization platform for supramolecular assemblies
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1LUH
 
 | | SOLUTION NMR STRUCTURE OF SELF-COMPLIMENTARY DUPLEX 5'-D(TCCG*CGGA)2 CONTAINING A TRIMETHYLENE CROSSLINK AT THE N2 POSITION OF G* | | Descriptor: | 5'-D(*TP*CP*CP*(TME)GP*CP*GP*GP*A)-3', PROPANE | | Authors: | Dooley, P.D, Zhang, M, Korbel, G.A, Nechev, L.V, Harris, C.M, Stone, M.P, Harris, T.M. | | Deposit date: | 2002-05-22 | | Release date: | 2003-02-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Determination of the Conformation of a Trimethylene Interstrand Cross-Link in an Oligodeoxynucleotide Duplex Containing a 5'-d(GpC) Motif
J.AM.CHEM.SOC., 125, 2003
|
|
