2O3J
 
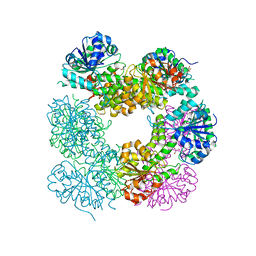 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
8S5N
 
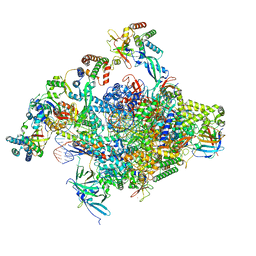 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 2024
|
|
8S55
 
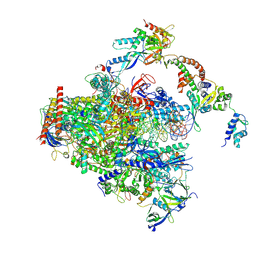 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 2024
|
|
8S51
 
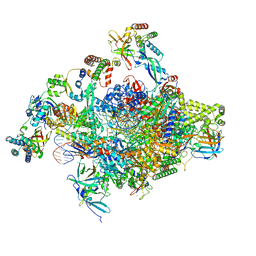 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 2024
|
|
8S52
 
 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 10 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 2024
|
|
8S54
 
 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 2024
|
|
1VZU
 
 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
1VZT
 
 | | ROLES OF INDIVIDUAL RESIDUES OF ALPHA-1,3 GALACTOSYLTRANSFERASES IN SUBSTRATE BINDING AND CATALYSIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, X, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-26 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
4OIP
 
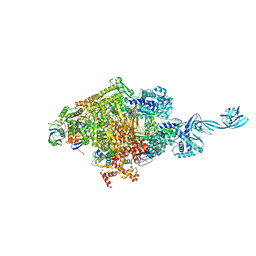 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
5UQ8
 
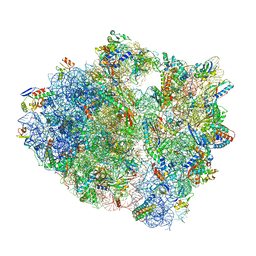 | | 70S ribosome complex with dnaX mRNA stem-loop and E-site tRNA ("out" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-03-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
4NPU
 
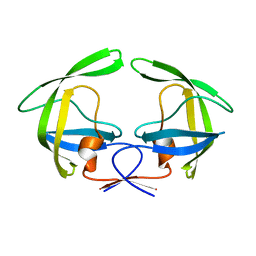 | |
3TKZ
 
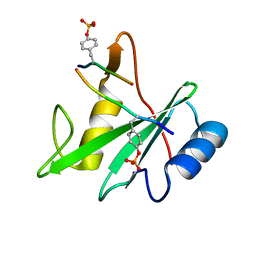 | | Structure of the SHP-2 N-SH2 domain in a 1:2 complex with RVIpYFVPLNR peptide | | Descriptor: | PROTEIN (RVIpYFVPLNR peptide), Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
3TL0
 
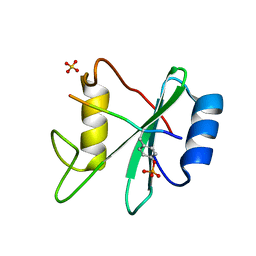 | | Structure of SHP2 N-SH2 domain in complex with RLNpYAQLWHR peptide | | Descriptor: | RLNpYAQLWHR peptide, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
8J57
 
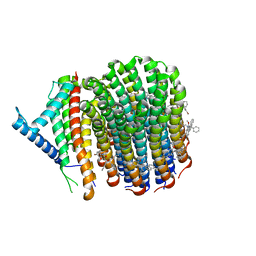 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in complex with bedaquiline(BDQ) | | Descriptor: | ATP synthase subunit a, ATP synthase subunit c, Bedaquiline | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure of Mycobacterium tuberculosis ATP synthase
To Be Published
|
|
8J58
 
 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in the apo-form | | Descriptor: | ATP synthase subunit a, ATP synthase subunit c | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure of Mycobacterium tuberculosis ATP synthase
To Be Published
|
|
8JR1
 
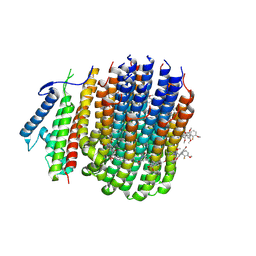 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in complex with TBAJ-587 | | Descriptor: | (1~{S},2~{S})-1-(6-bromanyl-2-methoxy-quinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2-fluoranyl-3-methoxy-phenyl)butan-2-ol, ATP synthase subunit a, ATP synthase subunit c | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of Mycobacterium tuberculosis ATP synthase
To Be Published
|
|
8JR0
 
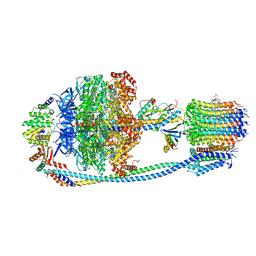 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase in complex with TBAJ-587 | | Descriptor: | (1~{S},2~{S})-1-(6-bromanyl-2-methoxy-quinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2-fluoranyl-3-methoxy-phenyl)butan-2-ol, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of Mycobacterium tuberculosis ATP synthase
To Be Published
|
|
6DS6
 
 | |
6O7G
 
 | |
3OMX
 
 | | Crystal structure of Ssu72 with vanadate complex | | Descriptor: | CG14216, VANADATE ION | | Authors: | Zhang, Y, Zhang, M, Zhang, Y. | | Deposit date: | 2010-08-27 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3366 Å) | | Cite: | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | Descriptor: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Zhang, Y, Hu, J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
7ST4
 
 | | Calcium-saturated jGCaMP8.410.80 | | Descriptor: | CALCIUM ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Zhang, Y, Looger, L.L. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fast and sensitive GCaMP calcium indicators for imaging neural populations
Nature, 615, 2023
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
2AA0
 
 | | Crystal structure of T. gondii adenosine kinase complexed with 6-methylmercaptopurine riboside | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2AB8
 
 | | Crystal structure of T. gondii adenosine kinase complexed with 6-methylmercaptopurine riboside and AMP-PCP | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
