6SHK
 
 | |
6Z4E
 
 | | The structure of the C-terminal domain of RssB from E. coli | | Descriptor: | Regulator of RpoS | | Authors: | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
7PDC
 
 | |
5NNT
 
 | | The dimeric structure of LL-37 crystallized in DPC | | Descriptor: | Cathelicidin antimicrobial peptide, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Zeth, K, Sancho-Vaello, E. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-24 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species.
Sci Rep, 7, 2017
|
|
6FHG
 
 | |
6ZLX
 
 | |
3H7Z
 
 | |
3H7X
 
 | |
6Z4C
 
 | | The structure of the N-terminal domain of RssB from E. coli | | Descriptor: | Regulator of RpoS | | Authors: | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
4N59
 
 | | The Crystal Structure of Pectocin M2 at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Pectocin M2, ... | | Authors: | Zeth, K, Grinter, R, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
3D9X
 
 | |
3LT6
 
 | |
3LT7
 
 | |
5NNK
 
 | | The structure of LL-37 crystallized in the presence LDAO | | Descriptor: | Cathelicidin antimicrobial peptide, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Zeth, K, Sancho-Vaello, E. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-24 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species.
Sci Rep, 7, 2017
|
|
5NMN
 
 | |
5NNM
 
 | | The crystal structure of dimeric LL-37 | | Descriptor: | CARBONATE ION, Cathelicidin antimicrobial peptide | | Authors: | Zeth, K, Sancho-Vaello, E. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-24 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species.
Sci Rep, 7, 2017
|
|
1TE0
 
 | |
4C4V
 
 | |
4ARQ
 
 | | Structure of the pesticin S89C, S285C double mutant | | Descriptor: | MAGNESIUM ION, PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARM
 
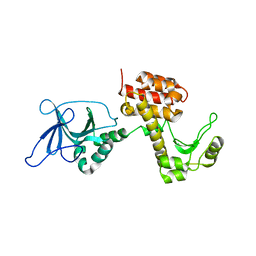 | | Structure of the inactive pesticin T201A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARP
 
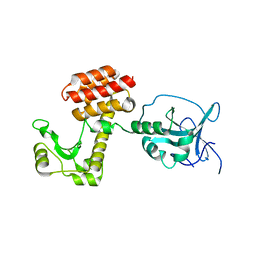 | | Structure of the inactive pesticin E178A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4AEQ
 
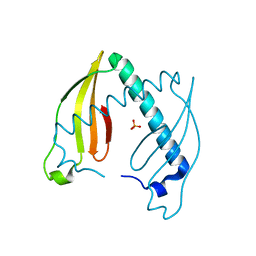 | | Crystal structure of the dimeric immunity protein Cmi solved by direct methods (Arcimboldo) | | Descriptor: | COLICIN-M IMMUNITY PROTEIN, PHOSPHATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Uson, I, Braun, V. | | Deposit date: | 2012-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | The Crystal Structure of the Dimeric Colicin M Immunity
J.Struct.Biol., 178, 2012
|
|
4ARJ
 
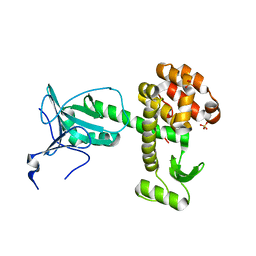 | | Crystal structure of a pesticin (translocation and receptor binding domain) from Y. pestis and T4-lysozyme chimera | | Descriptor: | PESTICIN, LYSOZYME, SULFATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-09 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARL
 
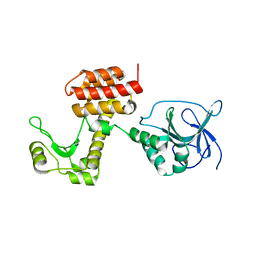 | | Structure of the inactive pesticin D207A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4CSO
 
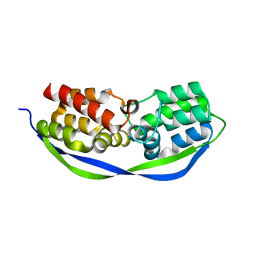 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
