8JME
 
 | |
8JMH
 
 | |
8JMI
 
 | |
8JMA
 
 | |
8JM9
 
 | |
6S8Z
 
 | |
8GWO
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWE
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 and GMPPNP | | 分子名称: | Helicase nsp13, MAGNESIUM ION, Non-structural protein 8, ... | | 著者 | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-16 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWB
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 | | 分子名称: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | 著者 | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-16 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWF
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.Y, Huang, Y.C, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWG
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWN
 
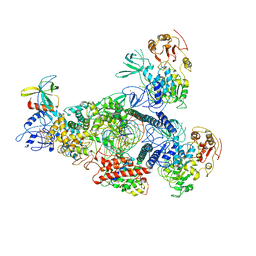 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWI
 
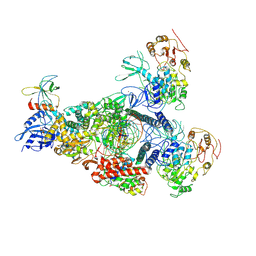 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GW1
 
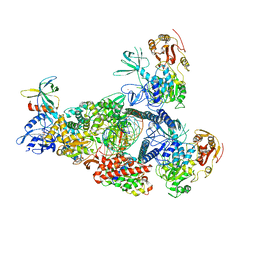 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | 分子名称: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | 著者 | Yan, L, Rao, Z, Lou, Z. | | 登録日 | 2022-09-16 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWM
 
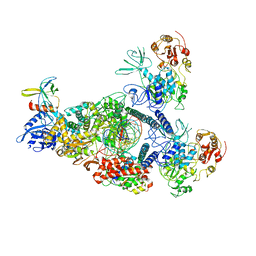 | | SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP | | 分子名称: | 2'-deoxy-2'-fluoro-2'-methyluridine 5'-(trihydrogen diphosphate), Helicase, Non-structural protein 7, ... | | 著者 | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWK
 
 | | SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP | | 分子名称: | Helicase, MAGNESIUM ION, Non-structural protein 7, ... | | 著者 | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2022-09-17 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
3PJZ
 
 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | 分子名称: | POTASSIUM ION, Potassium uptake protein TrkH | | 著者 | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2010-11-10 | | 公開日 | 2011-01-19 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.506 Å) | | 主引用文献 | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
8J17
 
 | | Crystal structure of IsPETase variant | | 分子名称: | Poly(ethylene terephthalate) hydrolase | | 著者 | Yin, Q.D, Wang, Y.X. | | 登録日 | 2023-04-12 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Efficient polyethylene terephthalate biodegradation by an engineered Ideonella sakaiensis PETase with a fixed substrate-binding W156 residue
Green Chem, 2013
|
|
5ZBH
 
 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | 分子名称: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | 著者 | Yang, Z, Han, S, Zhao, Q, Wu, B. | | 登録日 | 2018-02-11 | | 公開日 | 2018-04-25 | | 最終更新日 | 2018-05-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
5ZBQ
 
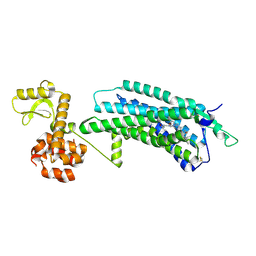 | | The Crystal Structure of human neuropeptide Y Y1 receptor with UR-MK299 | | 分子名称: | Neuropeptide Y receptor type 1,T4 Lysozyme, N~2~-(diphenylacetyl)-N-[(4-hydroxyphenyl)methyl]-N~5~-(N'-{[2-(propanoylamino)ethyl]carbamoyl}carbamimidoyl)-D-ornithinamide | | 著者 | Yang, Z, Han, S, Zhao, Q, Wu, B. | | 登録日 | 2018-02-12 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
8GU0
 
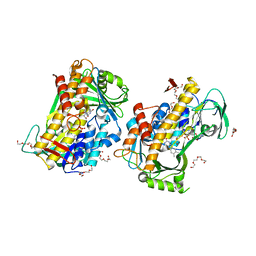 | |
8IAB
 
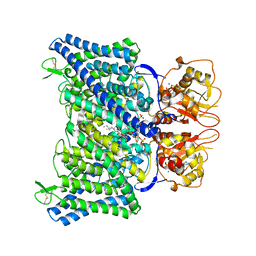 | | The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chloride channel protein CLC-a, ... | | 著者 | Yang, Z, Zhang, X, Zhang, P. | | 登録日 | 2023-02-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.96 Å) | | 主引用文献 | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8IAD
 
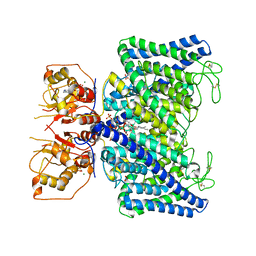 | | The Arabidopsis CLCa transporter bound with nitrate, ATP and PIP2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | 著者 | Yang, Z, Zhang, X, Zhang, P. | | 登録日 | 2023-02-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
1C78
 
 | | STAPHYLOKINASE (SAK) DIMER | | 分子名称: | STAPHYLOKINASE | | 著者 | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | 登録日 | 2000-02-01 | | 公開日 | 2000-08-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C77
 
 | | STAPHYLOKINASE (SAK) DIMER | | 分子名称: | STAPHYLOKINASE | | 著者 | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | 登録日 | 2000-02-01 | | 公開日 | 2000-08-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
