6L7I
 
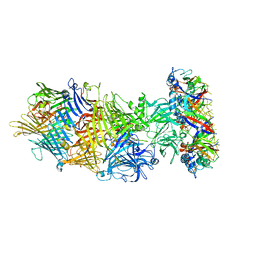 | |
6L7E
 
 | |
6L7A
 
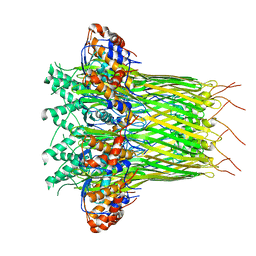 | | CsgFG complex in Curli biogenesis system | | Descriptor: | CsgF, Curli production assembly/transport protein CsgG | | Authors: | Yan, Z.F, Yin, M, Chen, J.N, Li, X.M. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Assembly and substrate recognition of curli biogenesis system.
Nat Commun, 11, 2020
|
|
6L7C
 
 | | CsgFG complex with substrate CsgAN6 peptide in Curli biogenesis system | | Descriptor: | CsgF, Curli production assembly/transport protein CsgG, Major curlin subunit CsgA | | Authors: | Yan, Z.F, Yin, M, Chen, J.N, Li, X.M. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Assembly and substrate recognition of curli biogenesis system.
Nat Commun, 11, 2020
|
|
7RXE
 
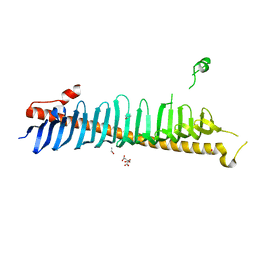 | | Crystal structure of junctophilin-2 | | Descriptor: | CITRATE ANION, ISOPROPYL ALCOHOL, Junctophilin-2 N-terminal fragment | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-22 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RW4
 
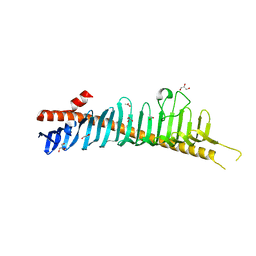 | | Crystal structure of junctophilin-1 | | Descriptor: | ACETATE ION, GLYCEROL, Junctophilin-1 | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RXQ
 
 | | Crystal structure of junctophilin-2 in complex with a CaV1.1 peptide | | Descriptor: | ETHANOL, Junctophilin-2 N-terminal fragment, SULFATE ION, ... | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-23 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9IUM
 
 | |
9IUK
 
 | | The structure of Candida albicans Cdr1 in apo state | | Descriptor: | Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-22 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUL
 
 | | The structure of Candida albicans Cdr1 in fluconazole-bound state | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
