6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
7ETW
 
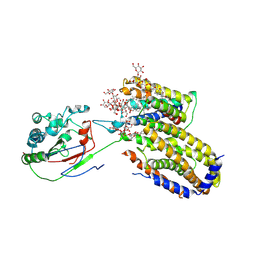 | | Cryo-EM structure of Scap/Insig complex in the present of digitonin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Digitonin, Insulin-induced gene 2 protein, ... | | Authors: | Yan, R, Cao, P, Song, W, Li, Y, Wang, T, Qian, H, Yan, C, Yan, N. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for sterol sensing by Scap and Insig
Cell Rep, 35, 2021
|
|
8AU1
 
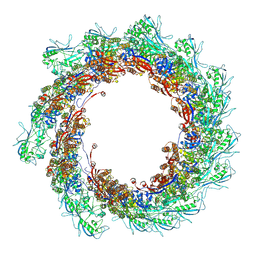 | | Jumbo Phage phi-kp24 tail outer sheath | | Descriptor: | Putative tail sheath protein | | Authors: | Ouyang, R, Briegel, A. | | Deposit date: | 2022-08-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8BFK
 
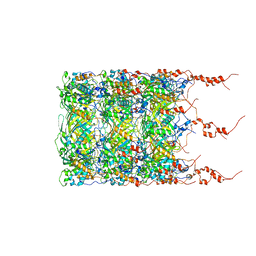 | | Jumbo Phage phi-kp24 tail inner tube | | Descriptor: | Putative virion structural protein | | Authors: | Ouyang, R, Briegel, A. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8BFL
 
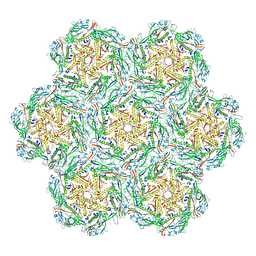 | | Jumbo Phage phi-kp24 empty capsid hexamers | | Descriptor: | Major head protein | | Authors: | Ouyang, R. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
8BFP
 
 | | Jumbo Phage phi-kp24 empty capsid pentamer hexamers | | Descriptor: | Major head protein | | Authors: | Ouyang, R. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
1JAC
 
 | | A NOVEL MODE OF CARBOHYDRATE RECOGNITION IN JACALIN, A MORACEAE PLANT LECTIN WITH A BETA-PRISM | | Descriptor: | JACALIN, methyl alpha-D-galactopyranoside | | Authors: | Sankaranarayanan, R, Sekar, K, Banerjee, R, Sharma, V, Surolia, A, Vijayan, M. | | Deposit date: | 1996-05-22 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a beta-prism fold.
Nat.Struct.Biol., 3, 1996
|
|
8F28
 
 | | Lysozyme Structures from Single-Entity Crystallization Method NanoAC | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yang, R, Sankaran, B, Ogbonna, E, Wang, G. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Single-Entity Method for Actively Controlled Nucleation and High-Quality Protein Crystal Synthesis.
Anal.Chem., 95, 2023
|
|
1EVL
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH A THREONYL ADENYLATE ANALOG | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1EVK
 
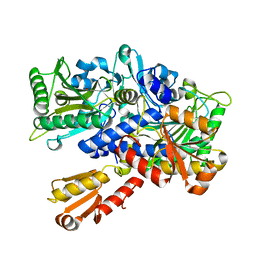 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH THE LIGAND THREONINE | | Descriptor: | THREONINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
3IT4
 
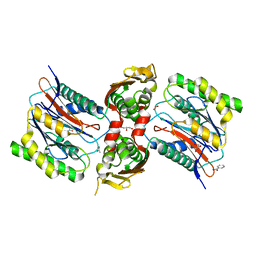 | | The Crystal Structure of Ornithine Acetyltransferase from Mycobacterium tuberculosis (Rv1653) at 1.7 A | | Descriptor: | ACETATE ION, Arginine biosynthesis bifunctional protein argJ alpha chain, Arginine biosynthesis bifunctional protein argJ beta chain, ... | | Authors: | Sankaranarayanan, R, Cherney, M.M, Garen, C, Garen, G, Yuan, M, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor.
J.Mol.Biol., 397, 2010
|
|
3IT6
 
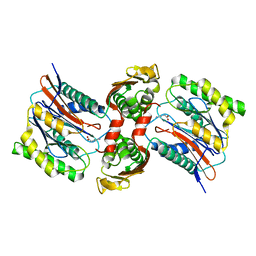 | | The Crystal Structure of Ornithine Acetyltransferase complexed with Ornithine from Mycobacterium tuberculosis (Rv1653) at 2.4 A | | Descriptor: | Arginine biosynthesis bifunctional protein argJ alpha chain, Arginine biosynthesis bifunctional protein argJ beta chain, L-ornithine | | Authors: | Sankaranarayanan, R, Cherney, M.M, Garen, C, Garen, G, Yuan, M, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor.
J.Mol.Biol., 397, 2010
|
|
1Y8I
 
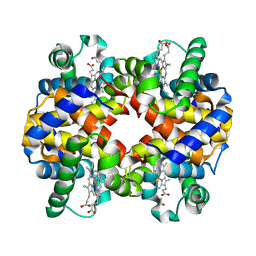 | | Horse methemoglobin low salt, PH 7.0 (98% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1Y8K
 
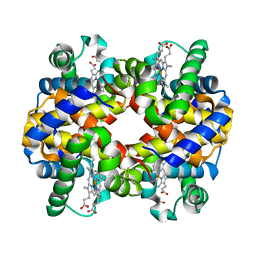 | | Horse methemoglobin low salt, PH 7.0 (88% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1Y8H
 
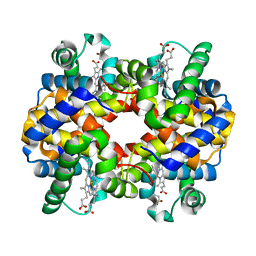 | | HORSE METHEMOGLOBIN LOW SALT, PH 7.0 | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
5H0I
 
 | |
1TEE
 
 | |
1TED
 
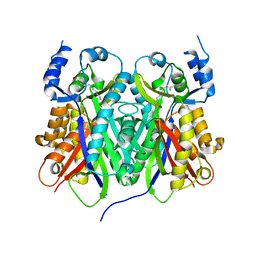 | |
1QF6
 
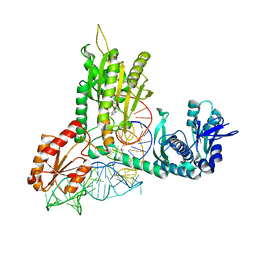 | | STRUCTURE OF E. COLI THREONYL-TRNA SYNTHETASE COMPLEXED WITH ITS COGNATE TRNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, THREONINE TRNA, THREONYL-TRNA SYNTHETASE, ... | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 1999-04-06 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site
Cell(Cambridge,Mass.), 97, 1999
|
|
8G5V
 
 | | Empty capsid of Hepatitis B virus | | Descriptor: | Core protein Cp183 | | Authors: | Yang, R, Cingolani, G. | | Deposit date: | 2023-02-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core.
Sci Adv, 10, 2024
|
|
8G6V
 
 | |
8G8Y
 
 | |
3AAX
 
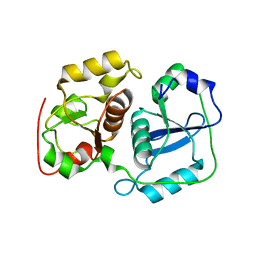 | | Crystal structure of probable thiosulfate sulfurtransferase cysa3 (RV3117) from Mycobacterium tuberculosis: monoclinic FORM | | Descriptor: | Putative thiosulfate sulfurtransferase | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
3AAY
 
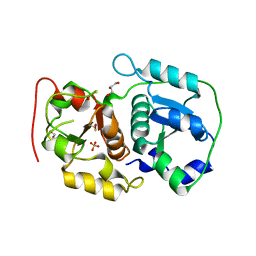 | | Crystal structure of probable thiosulfate sulfurtransferase CYSA3 (RV3117) from Mycobacterium tuberculosis: orthorhombic form | | Descriptor: | GLYCEROL, Putative thiosulfate sulfurtransferase, SULFATE ION | | Authors: | Sankaranarayanan, R, Witholt, S.J, Cherney, M.M, Garen, C.R, Cherney, L.T, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of probable thiosulfate sulfurtransferase CysA3 (Rv3117) from Mycobacterium tuberculosis
To be Published
|
|
6Y7C
 
 | | Early cytoplasmic yeast pre-40S particle (purified with Tsr1 as bait) | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Plassart, L, Plisson-Chastang, C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Good Vibrations: Structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy.
Molecules, 25, 2020
|
|
