7PDY
 
 | | A viral peptide from Marek's disease virus bound to chicken MHC-II molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 38 kDa phosphoprotein,MHC class II beta chain, ... | | Authors: | Goryanin, A, Cook, A.G, Kaufman, J, Halabi, S. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Viral peptide bound to chicken MHC-II molecule
To Be Published
|
|
5JZS
 
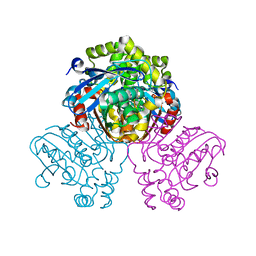 | | HsaD bound to 3,5-dichloro-4-hydroxybenzoic acid | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD, 3,5-dichloro-4-hydroxybenzoic acid | | Authors: | Ryan, A, Polycarpou, E, Lack, N, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E.D, Ballet, R, Abuhammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZB
 
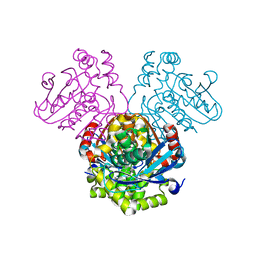 | | Crystal structure of HsaD bound to 3,5-dichlorobenzene sulphonamide | | Descriptor: | 3,5-dichlorobenzene-1-sulfonamide, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, PHOSPHATE ION | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZ9
 
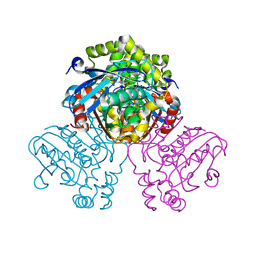 | | Crystal structure of HsaD bound to 3,5-dichloro-4-hydroxybenzenesulphonic acid | | Descriptor: | 3,5-dichloro-4-hydroxybenzene-1-sulfonic acid, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
8K4R
 
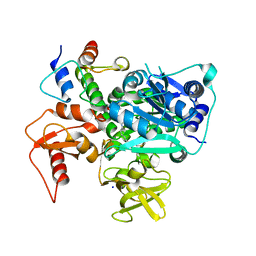 | | Structure of VinM-VinL complex | | Descriptor: | Acyl-carrier-protein, Non-ribosomal peptide synthetase, SODIUM ION, ... | | Authors: | Miyanaga, A, Nagata, K, Nakajima, J, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2023-07-20 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Amide-Forming Adenylation Enzyme VinM in Vicenistatin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
3LT5
 
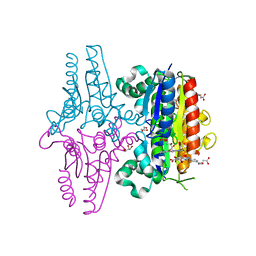 | | X-ray Crystallographic structure of a Pseudomonas Aeruginosa Azoreductase in complex with balsalazide | | Descriptor: | (3E)-3-({4-[(2-carboxyethyl)carbamoyl]phenyl}hydrazono)-6-oxocyclohexa-1,4-diene-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Ryan, A, Laurieri, N, Westwood, I, Wang, C.-J, Lowe, E, Sim, E. | | Deposit date: | 2010-02-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Azoreduction
J.Mol.Biol., 400, 2010
|
|
5Y1I
 
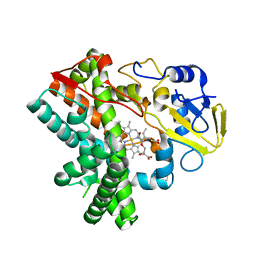 | | The crystal structure of GfsF | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2017-07-20 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Recognition by a Dual-Function P450 Monooxygenase GfsF Involved in FD-891 Biosynthesis
Chembiochem, 18, 2017
|
|
6C9Y
 
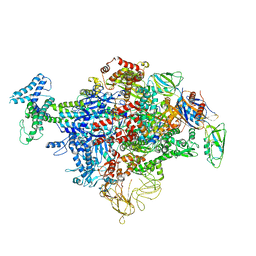 | | Cryo-EM structure of E. coli RNAP sigma70 holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Narayanan, A, Vago, F, Li, K, Qayyum, M.Z, Yenool, D, Jiang, W, Murakami, K.S. | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation.
J. Biol. Chem., 293, 2018
|
|
6CA0
 
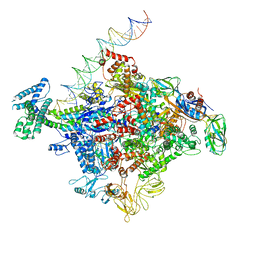 | | Cryo-EM structure of E. coli RNAP sigma70 open complex | | Descriptor: | DNA (35-MER), DNA (45-MER), DNA (5'-D(P*GP*CP*CP*GP*CP*GP*TP*CP*AP*GP*A)-3'), ... | | Authors: | Narayanan, A, Vago, F, Li, K, Qayyum, M.Z, Yernool, D, Jiang, W, Murakami, K.S. | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.75 Å) | | Cite: | Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation.
J. Biol. Chem., 293, 2018
|
|
7R2G
 
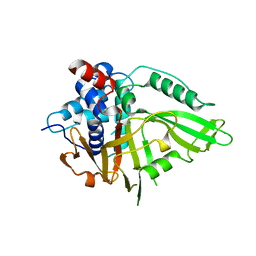 | | USP15 D1D2 in catalytically-competent state bound to mitoxantrone stack (isoform 2) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 15, ... | | Authors: | Priyanka, A, Sixma, T.K. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mitoxantrone stacking does not define the active or inactive state of USP15 catalytic domain.
J.Struct.Biol., 214, 2022
|
|
5CZD
 
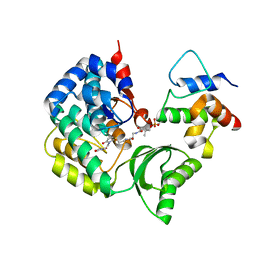 | | The complex structure of VinK with VinL | | Descriptor: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, Acyl-carrier-protein, ... | | Authors: | Miyanaga, A, Iwasawa, S, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CZC
 
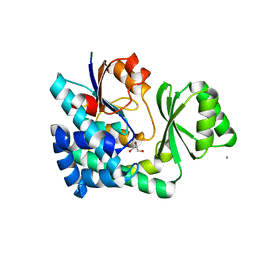 | | The structure of VinK | | Descriptor: | CALCIUM ION, GLYCEROL, Malonyl-CoA-[acyl-carrier-protein] transacylase | | Authors: | Miyanaga, A, Iwasawa, S, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3WFJ
 
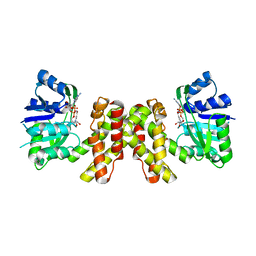 | | The complex structure of D-mandelate dehydrogenase with NADH | | Descriptor: | 2-dehydropantoate 2-reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
3WV5
 
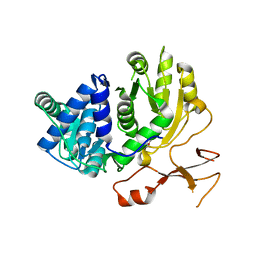 | | Complex structure of VinN with 3-methylaspartate | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WV4
 
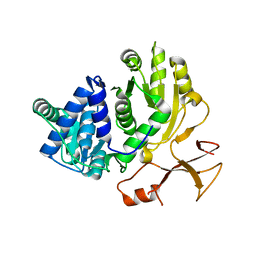 | | Crystal structure of VinN | | Descriptor: | Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WVN
 
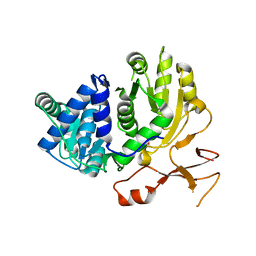 | | Complex structure of VinN with L-aspartate | | Descriptor: | ASPARTIC ACID, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-30 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WFI
 
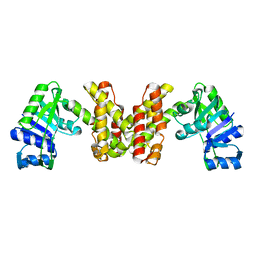 | | The crystal structure of D-mandelate dehydrogenase | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
2K7L
 
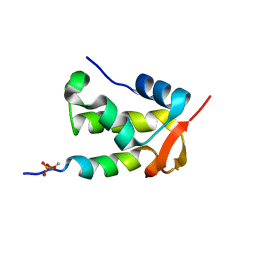 | | NMR structure of a complex formed by the C-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 | | Descriptor: | General transcription factor IIF subunit 1, centFCP1-T584PO4 peptide | | Authors: | Yang, A, Abbott, K.L, Desjardins, A, Di Lello, P, Omichinski, J.G, Legault, P. | | Deposit date: | 2008-08-13 | | Release date: | 2009-06-02 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex formed by the carboxyl-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 phosphatase
Biochemistry, 48, 2009
|
|
1IRE
 
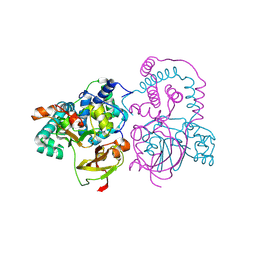 | | Crystal Structure of Co-type nitrile hydratase from Pseudonocardia thermophila | | Descriptor: | COBALT (II) ION, Nitrile Hydratase | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Wakagi, T. | | Deposit date: | 2001-10-01 | | Release date: | 2002-10-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cobalt-containing nitrile hydratase.
Biochem.Biophys.Res.Commun., 288, 2001
|
|
2A4J
 
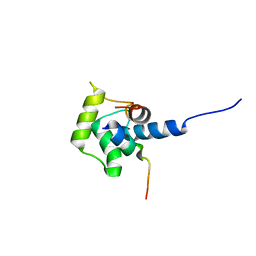 | | Solution structure of the C-terminal domain (T94-Y172) of the human centrin 2 in complex with a 17 residues peptide (P1-XPC) from xeroderma pigmentosum group C protein | | Descriptor: | 17-mer peptide P1-XPC from DNA-repair protein complementing XP-C cells, Centrin 2 | | Authors: | Yang, A, Miron, S, Mouawad, L, Duchambon, P, Blouquit, Y, Craescu, C.T. | | Deposit date: | 2005-06-29 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Flexibility and plasticity of human centrin 2 binding to the xeroderma pigmentosum group C protein (XPC) from nuclear excision repair.
Biochemistry, 45, 2006
|
|
3R6W
 
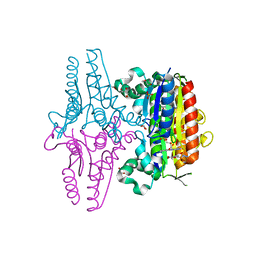 | | paAzoR1 binding to nitrofurazone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Ryan, A, Kaplan, K, Laurieri, N, Lowe, E, Sim, E. | | Deposit date: | 2011-03-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Activation of nitrofurazone by azoreductases: multiple activities in one enzyme.
Sci Rep, 1, 2011
|
|
1UGP
 
 | | Crystal structure of Co-type nitrile hydratase complexed with n-butyric acid | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGS
 
 | | Crystal structure of aY114T mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGQ
 
 | | Crystal structure of apoenzyme of Co-type nitrile hydratase | | Descriptor: | Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
1UGR
 
 | | Crystal structure of aT109S mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
