8I7L
 
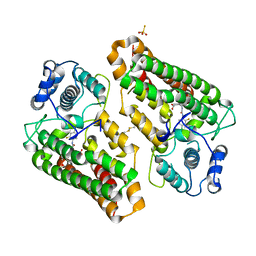 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with a novel inhibitor | | Descriptor: | 1-[3-[(4-chloranyl-2-fluoranyl-phenyl)carbamoylamino]-4-[cyclohexyl(2-methylpropyl)amino]phenyl]pyrrole-2-carboxylic acid, Indoleamine 2,3-dioxygenase 1, THIOSULFATE | | Authors: | Li, K, Liu, W, Dong, X. | | Deposit date: | 2023-02-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Apo-Form Selective Inhibition of IDO for Tumor Immunotherapy.
J Immunol., 209, 2022
|
|
8HF8
 
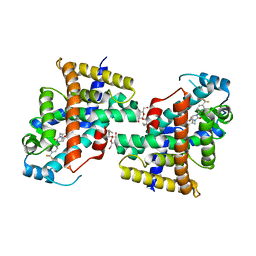 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist V1 | | Descriptor: | 2-[4-[[2,5-bis(oxidanylidene)-3-[4-(trifluoromethyl)phenyl]imidazolidin-1-yl]methyl]-2,6-dimethyl-phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of the First Subnanomolar PPAR alpha / delta Dual Agonist for the Treatment of Cholestatic Liver Diseases.
J.Med.Chem., 66, 2023
|
|
7XRS
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-05-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7XRY
 
 | | Crystal structure of MERS main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7YGQ
 
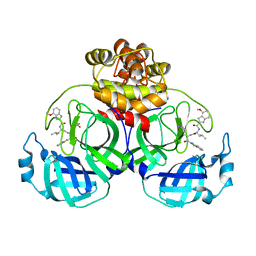 | | Crystal structure of SARS main protease in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7UVB
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT021601 | | Descriptor: | 2-hydroxy-6-({(3S)-4-[2-(2-hydroxyethyl)pyridine-3-carbonyl]morpholin-3-yl}methoxy)benzaldehyde, FORMYL GROUP, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Kaya, E, Xu, Q, Li, Z, Strutt, S.C, Cathers, B.E. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GBT021601 improves red blood cell health and the pathophysiology of sickle cell disease in a murine model.
Br.J.Haematol., 202, 2023
|
|
