8Z8Z
 
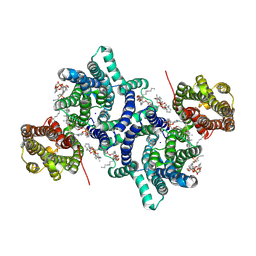 | | Cryo-EM structure of LYCHOS | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Xiong, Q, Zhu, Z, Li, T, Zhou, Z, Chao, Y, Qu, Q, Li, D. | | Deposit date: | 2024-04-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Molecular architecture of human LYCHOS involved in lysosomal cholesterol signaling.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7E2Q
 
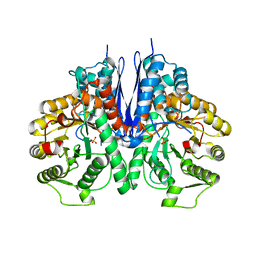 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7E2P
 
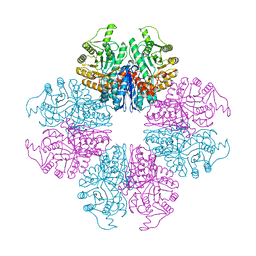 | | The Crystal Structure of Mycoplasma bovis enolase | | Descriptor: | Enolase | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Shao, G, Xiong, Q, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7U6R
 
 | |
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
6J36
 
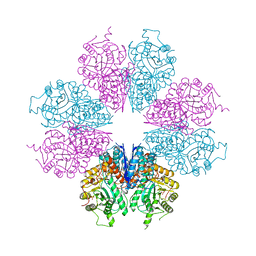 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
9E0I
 
 | |
9EH8
 
 | | Structure of the prefusion HKU5-19s Spike trimer (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Gen, R, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-11-22 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses.
Cell, 188, 2025
|
|
9EA0
 
 | | Structure of the prefusion HKU5-19s Spike trimer (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Gen, R, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-11-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses.
Cell, 188, 2025
|
|
8ZUF
 
 | | Cryo-EM structure of P.nat ACE2 mutant in complex with MOW15-22 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Tang, J, Deng, Z. | | Deposit date: | 2024-06-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses.
Cell, 188, 2025
|
|
9C45
 
 | |
9C44
 
 | |
9C6O
 
 | |
9D32
 
 | |
9DAK
 
 | |
7CYC
 
 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
7CYD
 
 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
