6JOY
 
 | | The X-ray Crystallographic Structure of Branching Enzyme from Rhodothermus obamensis STB05 | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB | | Authors: | Li, Z.F, Ban, X.F, Jiang, H.M, Wang, Z, Jin, T.C, Li, C.M, Gu, Z.B. | | Deposit date: | 2019-03-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Flexible Loop in Carbohydrate-Binding Module 48 Allosterically Modulates Substrate Binding of the 1,4-alpha-Glucan Branching Enzyme.
J.Agric.Food Chem., 69, 2021
|
|
8TMA
 
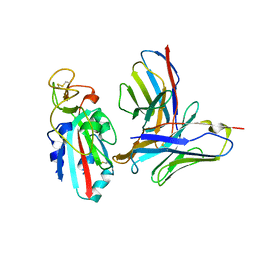 | |
8TM1
 
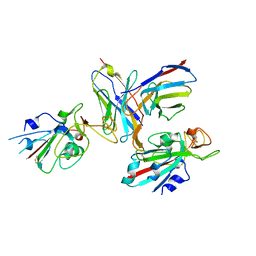 | |
8WD8
 
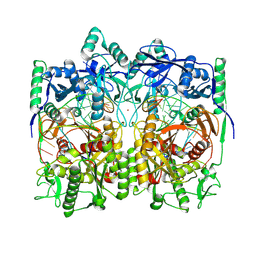 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-03-30 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
3O24
 
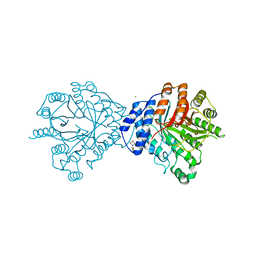 | | Crystal structure of the brevianamide F prenyltransferase FtmPT1 from Aspergillus fumigatus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, CHLORIDE ION, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
6LZE
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11a | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhang, Y, Jing, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
4E0U
 
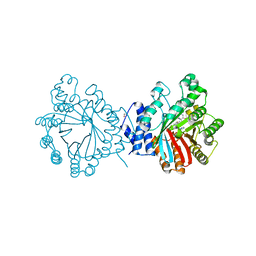 | | Crystal structure of CdpNPT in complex with thiolodiphosphate and (S)-benzodiazependione | | Descriptor: | (3S)-3-(1H-indol-3-ylmethyl)-3,4-dihydro-1H-1,4-benzodiazepine-2,5-dione, 1,2-ETHANEDIOL, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
4E0T
 
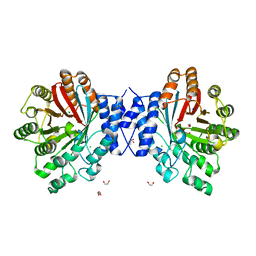 | | Crystal structure of CdpNPT in its unbound state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
4EJS
 
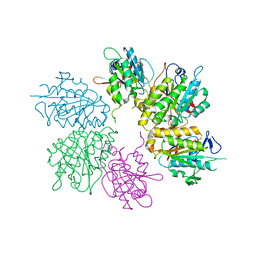 | | Structure of yeast elongator subcomplex Elp456 | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Lin, Z, Zhao, W, Long, J, Shen, Y. | | Deposit date: | 2012-04-07 | | Release date: | 2012-05-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure of elongator subcomplex Elp4-6
J.Biol.Chem., 287, 2012
|
|
5CQR
 
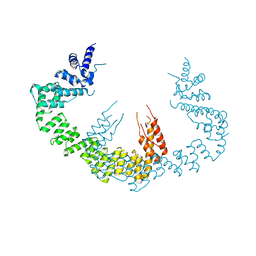 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CQS
 
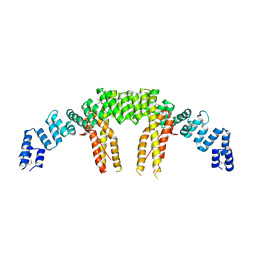 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7A3V
 
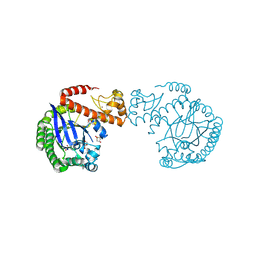 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with 3-hydroxysulfolane | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxythietane-1,1-dioxide, GLYCEROL, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
7A3X
 
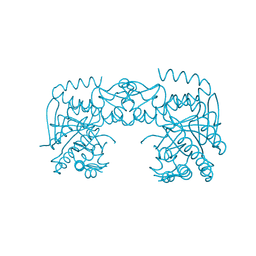 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with sulfolane | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-08-18 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
7A6D
 
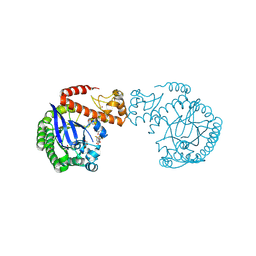 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with rac-trans-4,4-difluorocyclopentane-1,2-diol | | Descriptor: | (1~{R},2~{R})-4,4-bis(fluoranyl)cyclopentane-1,2-diol, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-08-25 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
7A4K
 
 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with tetramethylene sulfoxide | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-08-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
7A4X
 
 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with dimethyl sulfoxide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nguyen, D, Nguyen, T.X.P, Heine, A, Klebe, G. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
7ADN
 
 | | apo tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant | | Descriptor: | FORMIC ACID, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, D, You, S, Heine, A, Klebe, G. | | Deposit date: | 2020-09-15 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
7A0B
 
 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with rac-trans-3,4-dihydroxysulfolane | | Descriptor: | (3~{R},4~{R})-1,1-bis(oxidanylidene)thiolane-3,4-diol, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-08-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
7A9E
 
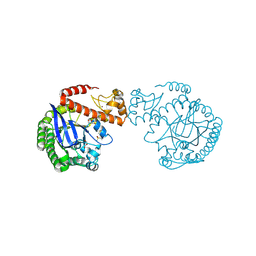 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with ethyl methyl sulfone | | Descriptor: | 1,2-ETHANEDIOL, 1-methylsulfonylethane, GLYCEROL, ... | | Authors: | Nguyen, D, You, S, Heine, A, Klebe, G. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
6LI1
 
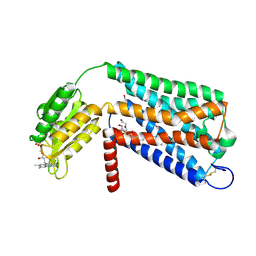 | | Crystal structure of GPR52 ligand free form with flavodoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Flavodoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI2
 
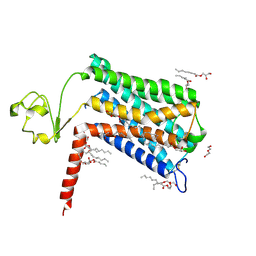 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
