6KKN
 
 | | Crystal structure of RuBisCO accumulation factor Raf1 from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-07-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6KKM
 
 | | Crystal structure of RbcL-Raf1 complex from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein, Ribulose bisphosphate carboxylase large chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-07-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6LRR
 
 | | Cryo-EM structure of RuBisCO-Raf1 from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6LRS
 
 | | Cryo-EM structure of RbcL8-RbcS4 from Anabaena sp. PCC 7120 | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Sun, H, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-01-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
7XSD
 
 | |
7D6C
 
 | | Crystal structure of CcmM N-terminal domain in complex with CcmN | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Carboxysome assembly protein CcmN | | Authors: | Sun, H, Cui, N, Han, S.J, Chen, Z.P, Xia, L.Y, Chen, Y, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Complex structure reveals CcmM and CcmN form a heterotrimeric adaptor in beta-carboxysome.
Protein Sci., 30, 2021
|
|
8Y1B
 
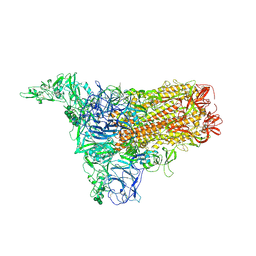 | | 1up-2 conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1H
 
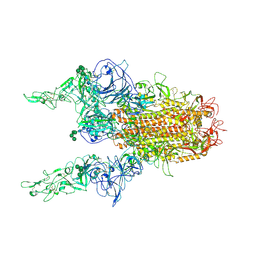 | | The 2up formation of the HKU1-B S protein in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1E
 
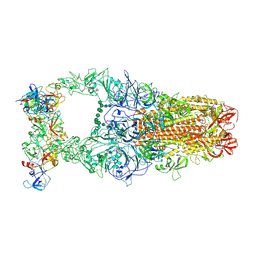 | | 3up-TM conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1G
 
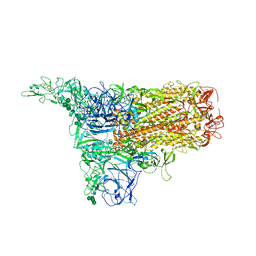 | | The 1up conformation of the HKU1-B S protein in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1C
 
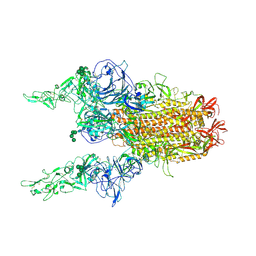 | | 2up-1 conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1A
 
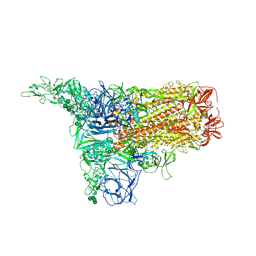 | | 1up-1 conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y19
 
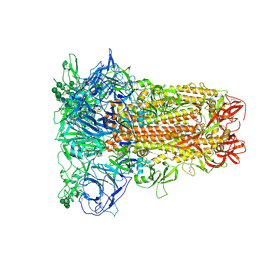 | | Closed conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1F
 
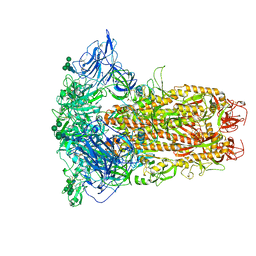 | | The closed conformation of the HKU1-B S protein in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1D
 
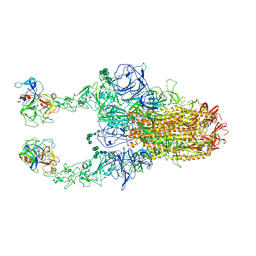 | | 2up-TM conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
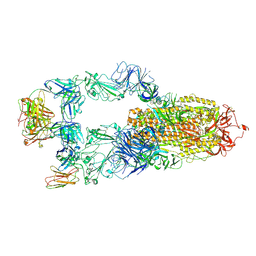
8YWX
 
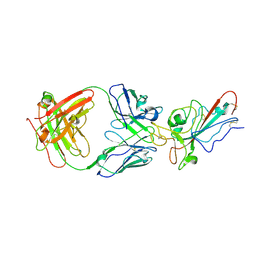 | |
8YWW
 
 | | The structure of HKU1-B S protein with bsAb1 | | Descriptor: | H4B6 heavy chain, H4B6 light chain, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-04-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A novel bispecific antibody targeting two overlapping epitopes in RBD improves neutralizing potency and breadth against SARS-CoV-2.
Emerg Microbes Infect, 13, 2024
|
|
5Z73
 
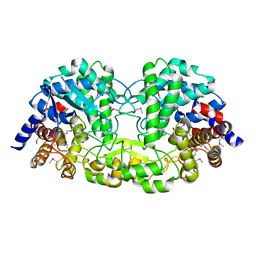 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 | | Descriptor: | Alr0819 protein, GLYCEROL | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
5Z74
 
 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alr0819 protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
