2GA7
 
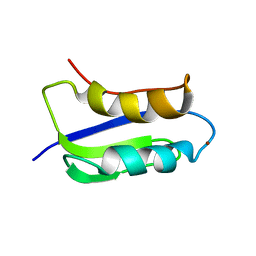 | | Solution structure of the copper(I) form of the third metal-binding domain of ATP7A protein (menkes disease protein) | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-01 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2GRI
 
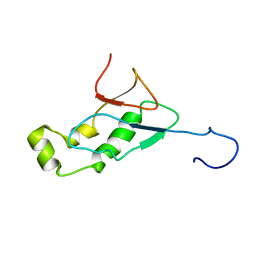 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2I3B
 
 | |
2KC6
 
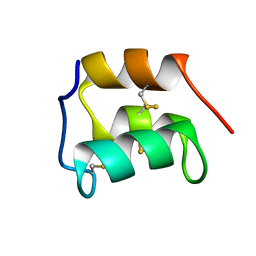 | | NMR solution structure of the pheromone En-1 of Euplotes nobilii at -1.5 C | | Descriptor: | Mating pheromone En-1 | | Authors: | Pedrini, B, Alimenti, C, Vallesi, A, Luporini, P, Wuthrich, K. | | Deposit date: | 2008-12-17 | | Release date: | 2009-08-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Molecular cold-adaptation: Comparative analysis of two homologous families of psychrophilic and mesophilic signal proteins of the protozoan ciliate, Euplotes.
Iubmb Life, 61, 2009
|
|
2LLG
 
 | | NMR structure of the protein NP_814968.1 from Enterococcus faecalis | | Descriptor: | Uncharacterized protein | | Authors: | Susac, L, Serrano, P, Geralt, M, Mohanty, B, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_814968.1 from Enterococcus faecalis
To be Published
|
|
2LYY
 
 | | NMR structure of the protein NB7890A from Shewanella sp | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Pedrini, B, Wuthrich, K, Horst, R, Augustyniak, W, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NB7890A from Shewanella sp
To be Published
|
|
2MHN
 
 | |
2M2B
 
 | | NMR structure of the RRM2 domain of the protein RBM10 from Homo sapiens | | Descriptor: | RNA-binding protein 10 | | Authors: | Serrano, P, Geralt, M, Dutta, S.K, Wuthrich, K, Wrobel, R.L, Makino, S, Misenhiemer, T.M, Markley, J.L, Fox, B.G, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the RRM2 domain of the protein RBM10 from Homo sapiens
To be Published
|
|
2M7O
 
 | |
2MQB
 
 | |
2MHL
 
 | |
2MDZ
 
 | | NMR structure of the Paracoccus denitrificans Z-subunit determined in the presence of ADP | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Wuthrich, K, Morales-Rios, E, Zarco-Zavala, M, Garcia-Trejo, J.J, Dutta, S.K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans
To be Published
|
|
2N6D
 
 | |
2MXT
 
 | |
2NBB
 
 | |
2N8G
 
 | | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens | | Descriptor: | Homeobox protein GBX-1 | | Authors: | Proudfoot, A.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens
To be Published
|
|
2N6E
 
 | |
2NSW
 
 | |
2NSV
 
 | |
4MT2
 
 | |
3GJO
 
 | |
1ADZ
 
 | |
1BYX
 
 | | CHIMERIC HYBRID DUPLEX R(GCAGUGGC).R(GCCA)D(CTGC) COMPRISING THE TRNA-DNA JUNCTION FORMED DURING INITIATION OF HIV-1 REVERSE TRANSCRIPTION | | Descriptor: | DNA/RNA (5'-R(*GP*CP*CP*A)-D(P*CP*TP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*UP*GP*GP*C)-3') | | Authors: | Szyperski, T, Goette, M, Billeter, M, Perola, E, Cellai, L. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the chimeric hybrid duplex r(gcaguggc).r(gcca)d(CTGC) comprising the tRNA-DNA junction formed during initiation of HIV-1 reverse transcription.
J.Biomol.NMR, 13, 1999
|
|
5CXT
 
 | |
1CXR
 
 | |
