5JP4
 
 | |
7PMM
 
 | |
7PMQ
 
 | |
7BBB
 
 | |
7PLI
 
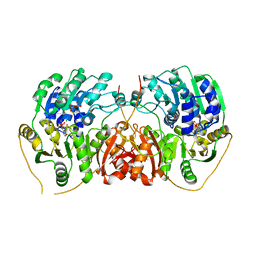 | |
5APG
 
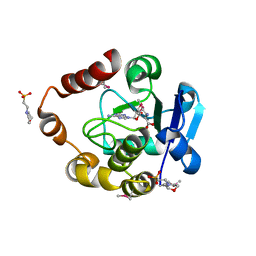 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | 著者 | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | 登録日 | 2015-09-15 | | 公開日 | 2016-04-27 | | 最終更新日 | 2016-06-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
5AP8
 
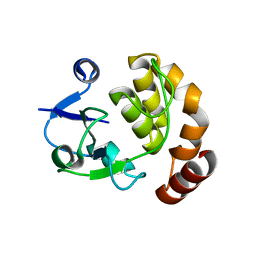 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | 分子名称: | TSR3 | | 著者 | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | 登録日 | 2015-09-14 | | 公開日 | 2016-04-27 | | 最終更新日 | 2016-06-01 | | 実験手法 | X-RAY DIFFRACTION (2.246 Å) | | 主引用文献 | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
2LCQ
 
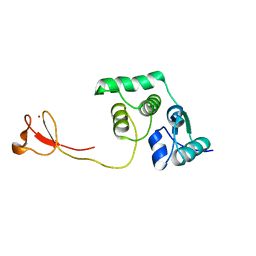 | | Solution structure of the endonuclease Nob1 from P.horikoshii | | 分子名称: | Putative toxin VapC6, ZINC ION | | 著者 | Veith, T, Martin, R, Wurm, J.P, Weis, B, Duchardt-Ferner, E, Safferthal, C, Hennig, R, Mirus, O, Bohnsack, M.T, Woehnert, J, Schleiff, E. | | 登録日 | 2011-05-05 | | 公開日 | 2011-12-14 | | 最終更新日 | 2012-05-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and functional analysis of the archaeal endonuclease Nob1.
Nucleic Acids Res., 40, 2012
|
|
7PVM
 
 | |
7AC8
 
 | |
5N2V
 
 | |
4QVK
 
 | | Apo-crystal structure of Podospora anserina methyltransferase PaMTH1 | | 分子名称: | 1,2-ETHANEDIOL, PaMTH1 Methyltransferase | | 著者 | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | 登録日 | 2014-07-15 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
6TRQ
 
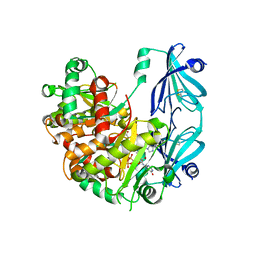 | | S.c. Scavenger Decapping Enzyme DcpS in complex with the capped RNA dinucleotide m7G-GU | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, L-GUANOSINE-5'-MONOPHOSPHATE, PHOSPHONATE, ... | | 著者 | Fuchs, A.-L, Neu, A, Sprangers, R. | | 登録日 | 2019-12-19 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.944 Å) | | 主引用文献 | Molecular basis of the selective processing of short mRNA substrates by the DcpS mRNA decapping enzyme.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7OPK
 
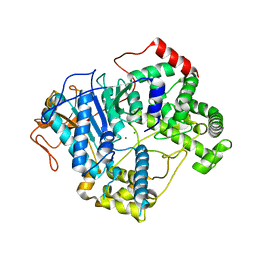 | |
2N0J
 
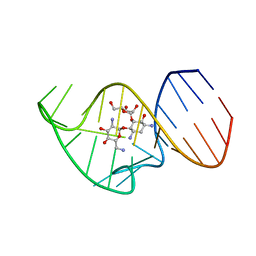 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | 分子名称: | RIBOSTAMYCIN, RNA_(27-MER) | | 著者 | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | 登録日 | 2015-03-09 | | 公開日 | 2016-02-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6HAG
 
 | |
6HTJ
 
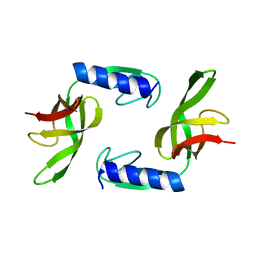 | |
4YMG
 
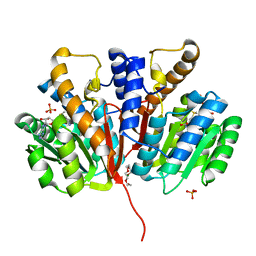 | | Crystal structure of SAM-bound Podospora anserina methyltransferase PaMTH1 | | 分子名称: | MAGNESIUM ION, PHOSPHATE ION, Putative SAM-dependent O-methyltranferase, ... | | 著者 | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | 登録日 | 2015-03-06 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.899 Å) | | 主引用文献 | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
4YMH
 
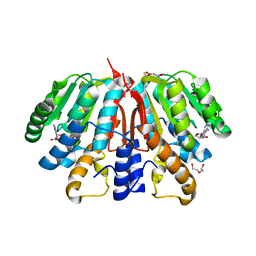 | | Crystal structure of SAH-bound Podospora anserina methyltransferase PaMTH1 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Putative SAM-dependent O-methyltranferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | 登録日 | 2015-03-06 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.876 Å) | | 主引用文献 | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
4I68
 
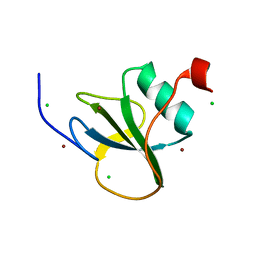 | |
4I67
 
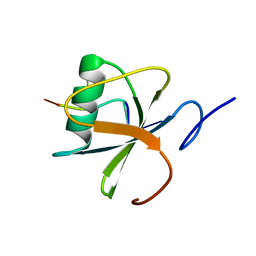 | |
4I69
 
 | |
2MXS
 
 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | 分子名称: | PAROMOMYCIN, RNA (27-MER) | | 著者 | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | 登録日 | 2015-01-14 | | 公開日 | 2015-12-09 | | 最終更新日 | 2016-02-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6EWT
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | 分子名称: | NRPS Kj12B-NDD | | 著者 | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | 登録日 | 2017-11-06 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWV
 
 | | Solution Structure of Docking Domain Complex of RXP NRPS: Kj12C NDD - Kj12B CDD | | 分子名称: | NRPS Kj12C-NDD, NRPS Kj12B-CDD | | 著者 | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | 登録日 | 2017-11-06 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
