5RVC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000933940912 | | 分子名称: | (1R,5R)-N-methyl-N-(1H-pyrazol-4-yl)bicyclo[3.1.0]hexane-1-carboxamide, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVT
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002582714 | | 分子名称: | 6-methyl-1H-indole-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-10-02 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSK
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000901381520_N3 | | 分子名称: | 3-[(3-methoxy-1,2-oxazol-5-yl)methyl]-3H-purin-6-amine, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSZ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000004218283 | | 分子名称: | 1-PHENYL-1H-PYRAZOLE-4-CARBOXYLIC ACID, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTE
 
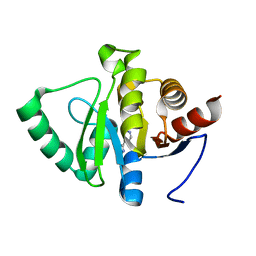 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000013283576 | | 分子名称: | 4-(1H-imidazol-2-yl)pyridine, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTV
 
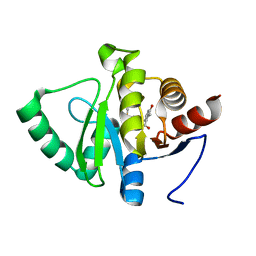 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001698894 | | 分子名称: | 3-hydroxy-2-methylbenzoic acid, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUS
 
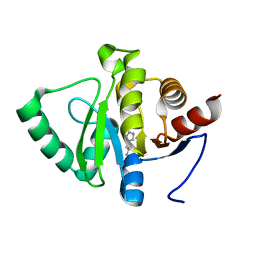 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388081 | | 分子名称: | HISTAMINE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RV9
 
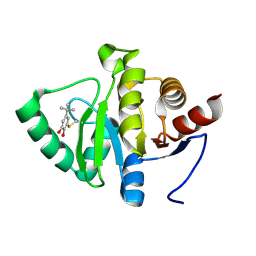 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388150 | | 分子名称: | 4-tert-butylbenzene-1,2-diol, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVO
 
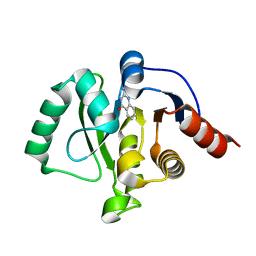 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000013514509 | | 分子名称: | 2-AMINOQUINAZOLIN-4(3H)-ONE, Non-structural protein 3 | | 著者 | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-10-02 | | 公開日 | 2020-12-16 | | 最終更新日 | 2022-05-25 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6LQA
 
 | | voltage-gated sodium channel Nav1.5 with quinidine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Quinidine, Sodium channel protein type 5 subunit alpha | | 著者 | Yan, N, Li, Z, Pan, X, Huang, G. | | 登録日 | 2020-01-13 | | 公開日 | 2021-03-24 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural Basis for Pore Blockade of the Human Cardiac Sodium Channel Na v 1.5 by the Antiarrhythmic Drug Quinidine*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4UMT
 
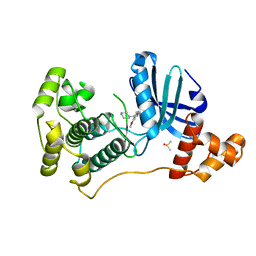 | | Structure of MELK in complex with inhibitors | | 分子名称: | 1-(4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}-2-methoxybenzyl)piperazinediium, DIMETHYL SULFOXIDE, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | 著者 | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | 登録日 | 2014-05-21 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMU
 
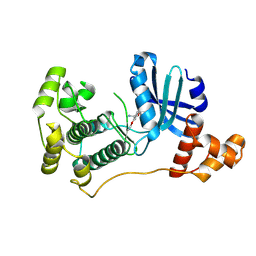 | | Structure of MELK in complex with inhibitors | | 分子名称: | (2-ethoxy-4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}phenyl)methanaminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | 著者 | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | 登録日 | 2014-05-21 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
1Q6B
 
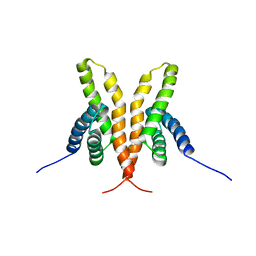 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Ensemble of 25 Structures | | 分子名称: | Circadian clock protein KaiA homolog | | 著者 | Vakonakis, I, Sun, J, Golden, S.S, Holzenburg, A, LiWang, A.C. | | 登録日 | 2003-08-13 | | 公開日 | 2003-08-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q6A
 
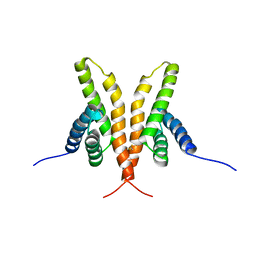 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Averaged Minimized Structure | | 分子名称: | Circadian clock protein KaiA homolog | | 著者 | Vakonakis, I, Sun, J, Holzenburg, A, Golden, S.S, LiWang, A.C. | | 登録日 | 2003-08-13 | | 公開日 | 2003-08-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Li, S, Zheng, Q. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-08-11 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Zheng, Q, Li, S. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-08-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
7F17
 
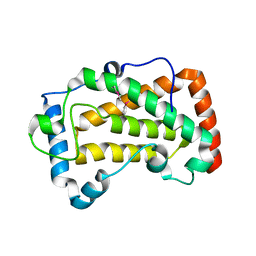 | | Crystal Structure of acid phosphatase | | 分子名称: | Acid phosphatase | | 著者 | Xu, X, Hou, X.D, Song, W, Rao, Y.J, Liu, L.M, Wu, J. | | 登録日 | 2021-06-08 | | 公開日 | 2021-10-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
2PL9
 
 | |
2PMC
 
 | |
4DK5
 
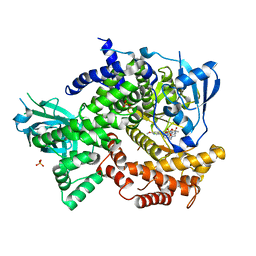 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine inhibitor | | 分子名称: | 4-(2-[(6-methoxypyridin-3-yl)amino]-5-{[4-(methylsulfonyl)piperazin-1-yl]methyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2012-02-03 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure-based design of a novel series of potent, selective inhibitors of the class I phosphatidylinositol 3-kinases.
J.Med.Chem., 55, 2012
|
|
3LRU
 
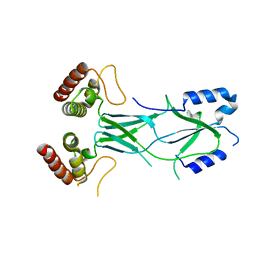 | |
7F18
 
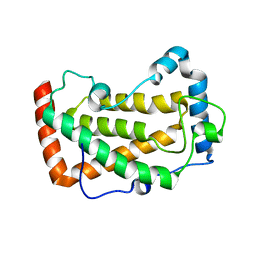 | | Crystal Structure of a mutant of acid phosphatase from Pseudomonas aeruginosa (Q57H/W58P/D135R) | | 分子名称: | Acid phosphatase | | 著者 | Xu, X, Hou, X.D, Song, W, Yin, D.J, Rao, Y.J, Liu, L.M. | | 登録日 | 2021-06-08 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
6IMA
 
 | | Crystal Structure of ALKBH1 without alpha-1 (N37-C369) | | 分子名称: | CITRIC ACID, Nucleic acid dioxygenase ALKBH1 | | 著者 | Zhang, M, Yang, S, Zhao, W, Li, H. | | 登録日 | 2018-10-22 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-04-08 | | 実験手法 | X-RAY DIFFRACTION (2.593 Å) | | 主引用文献 | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
6IMC
 
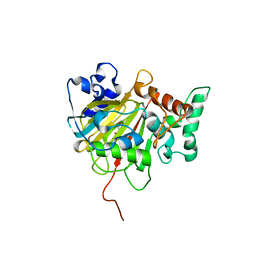 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | 分子名称: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | 著者 | Zhang, M, Yang, S, Zhao, W, Li, H. | | 登録日 | 2018-10-22 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
7EK6
 
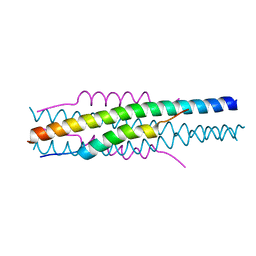 | | Structure of viral peptides IPB19/N52 | | 分子名称: | Spike protein S2 | | 著者 | Yu, D, Qin, B, Cui, S, He, Y. | | 登録日 | 2021-04-04 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.243 Å) | | 主引用文献 | Structure-based design and characterization of novel fusion-inhibitory lipopeptides against SARS-CoV-2 and emerging variants.
Emerg Microbes Infect, 10, 2021
|
|
