8KI5
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, PhmA | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
8KIH
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, diterpene synthase, ... | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
6B0D
 
 | | An E. coli DPS protein from ferritin superfamily | | Descriptor: | DNA protection during starvation protein, FORMIC ACID, SODIUM ION | | Authors: | Rui, W, Ruslan, S, Ronan, K, Adam, J.S. | | Deposit date: | 2017-09-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SIMBAD: a sequence-independent molecular-replacement pipeline.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6B6M
 
 | | Cyanase from Serratia proteamaculans | | Descriptor: | Cyanate hydratase | | Authors: | Xu, Y. | | Deposit date: | 2017-10-02 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SIMBAD: a sequence-independent molecular-replacement pipeline.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6BY0
 
 | |
6K0Y
 
 | | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Liu, J.X, Wang, G.Q. | | Deposit date: | 2019-05-08 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Sci Rep, 9, 2019
|
|
6KJZ
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 1 | | Descriptor: | 5,9-dihydroxy-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-en-1-yl)-3,4-dihydro-2H,6H-pyrano[3,2-b]xanthen-6-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2019-07-23 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.200001 Å) | | Cite: | Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
J.Med.Chem., 63, 2020
|
|
6KK0
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 4e | | Descriptor: | 7-[8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-5-oxidanyl-6-oxidanylidene-pyrano[3,2-b]xanthen-9-yl]oxyheptanoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2019-07-23 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.20008755 Å) | | Cite: | Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
J.Med.Chem., 63, 2020
|
|
7Y9G
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7TZP
 
 | | Crystal Structure of Putataive Short-Chain Dehydrogenase/Reductase (FabG) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 in Complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-ACP reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-02 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6JRK
 
 | | The structure of co-crystals of 8r-B-EGFR WT complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6JRJ
 
 | | The structure of co-crystals of 8r-B-EGFR T790M/C797S complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.943 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
3GRF
 
 | |
3OON
 
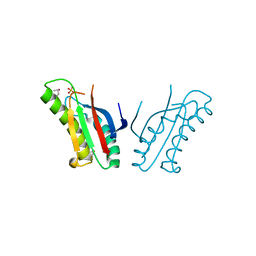 | | The structure of an outer membrance protein from Borrelia burgdorferi B31 | | Descriptor: | Outer membrane protein (Tpn50), SULFATE ION | | Authors: | Fan, Y, Bigelow, L, Feldman, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
3KZF
 
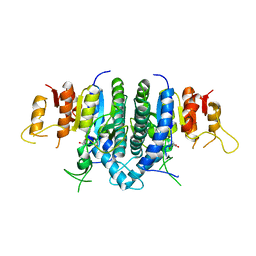 | | Structure of Giardia Carbamate Kinase | | Descriptor: | Carbamate kinase, GLYCEROL | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-12-08 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure and characterization of carbamate kinase from the human parasite Giardia lamblia.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2LE8
 
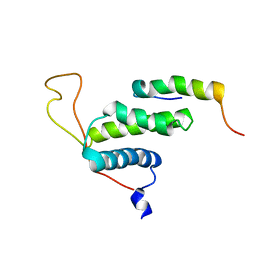 | | The protein complex for DNA replication | | Descriptor: | DNA replication factor Cdt1, DNA replication licensing factor MCM6 | | Authors: | Liu, C, Wei, Z, Zhu, G. | | Deposit date: | 2011-06-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the Cdt1-mediated MCM2-7 chromatin loading
Nucleic Acids Res., 40, 2012
|
|
5XG8
 
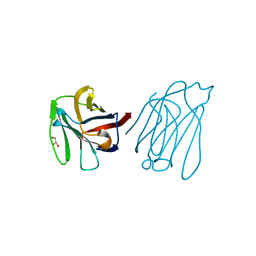 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | GLYCEROL, Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J.Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
5XRG
 
 | | Galectin-10/Charcot-Leyden crystal protein crystal structure | | Descriptor: | GLYCEROL, Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
5XRK
 
 | |
5XRP
 
 | | Galectin-10/Charcot-Leyden crystal protein variant Q75A | | Descriptor: | GLYCEROL, Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
5XRI
 
 | |
5XRO
 
 | |
5XG7
 
 | | Galectin-13/Placental Protein 13 crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y, Wang, Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
