6ZYD
 
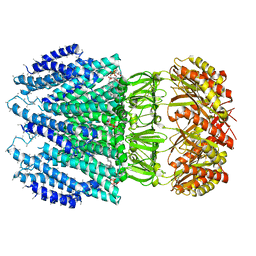 | | YnaI | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI,Low conductance mechanosensitive channel YnaI | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-07-31 | | Release date: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1R5P
 
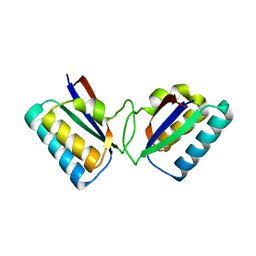 | |
1R5Q
 
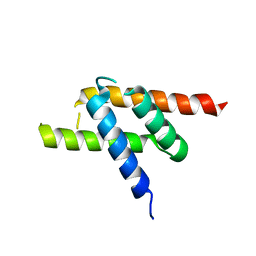 | |
1YTV
 
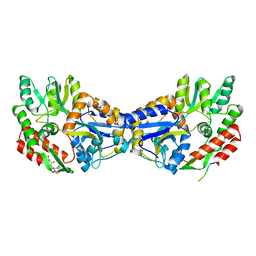 | | Maltose-binding protein fusion to a C-terminal fragment of the V1a vasopressin receptor | | Descriptor: | Maltose-binding periplasmic protein, Vasopressin V1a receptor, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Adikesavan, N.V, Mahmood, S.S, Stanley, S, Xu, Z, Wu, N, Thibonnier, M, Shoham, M. | | Deposit date: | 2005-02-11 | | Release date: | 2005-04-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-terminal segment of the V1R vasopressin receptor is unstructured in the crystal structure of its chimera with the maltose-binding protein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1JCH
 
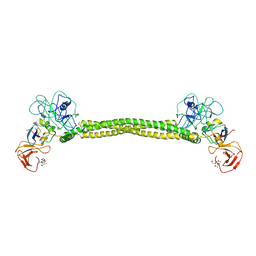 | | Crystal Structure of Colicin E3 in Complex with its Immunity Protein | | Descriptor: | CITRIC ACID, COLICIN E3, COLICIN E3 IMMUNITY PROTEIN, ... | | Authors: | Soelaiman, S, Jakes, K, Wu, N, Li, C, Shoham, M. | | Deposit date: | 2001-06-09 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of colicin E3: implications for cell entry and ribosome inactivation.
Mol.Cell, 8, 2001
|
|
1YRK
 
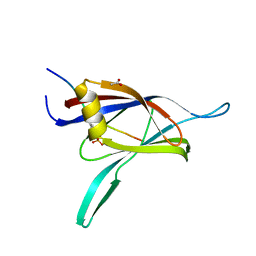 | | The C2 Domain of PKC is a new Phospho-Tyrosine Binding Domain | | Descriptor: | 13-residue peptide, ACETIC ACID, Protein kinase C, ... | | Authors: | Benes, C.H, Wu, N, Elia, A.E, Dharia, T, Cantley, L.C, Soltoff, S.P. | | Deposit date: | 2005-02-03 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C2 domain of PKCdelta is a phosphotyrosine binding domain.
Cell(Cambridge,Mass.), 121, 2005
|
|
2GL2
 
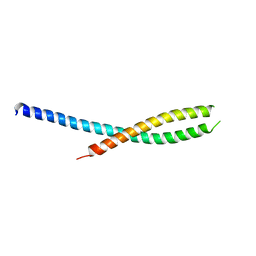 | | Crystal structure of the tetra mutant (T66G,R67G,F68G,Y69G) of bacterial adhesin FadA | | Descriptor: | adhesion A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2006-04-04 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary X-ray data of the FadA adhesin from Fusobacterium nucleatum.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1T6H
 
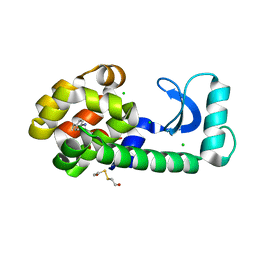 | | Crystal Structure T4 Lysozyme incorporating an unnatural amino acid p-iodo-L-phenylalanine at position 153 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Spraggon, G, Xie, J, Wang, L, Wu, N, Brock, A, Schultz, P.G. | | Deposit date: | 2004-05-06 | | Release date: | 2004-10-26 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination.
Nat.Biotechnol., 22, 2004
|
|
3ETW
 
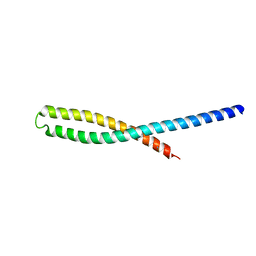 | | Crystal Structure of bacterial adhesin FadA | | Descriptor: | Adhesin A, THIOCYANATE ION | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETY
 
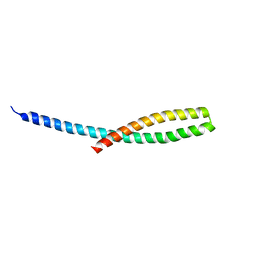 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETX
 
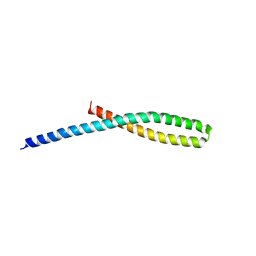 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETZ
 
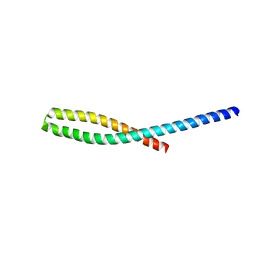 | | Crystal structure of bacterial adhesin FadA L76A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
7N82
 
 | | NMR Solution structure of Se0862 | | Descriptor: | Biofilm-related protein | | Authors: | Zhang, N, LiWang, A.L. | | Deposit date: | 2021-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2.
Proteins, 89, 2021
|
|
4UF0
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with epitherapuetic compound 2-(((2-((2-(dimethylamino)ethyl) (ethyl)amino)-2-oxoethyl)amino)methyl)isonicotinic acid. | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, FE (II) ION, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Tobias, K, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5A1F
 
 | | Crystal structure of the catalytic domain of PLU1 in complex with N-oxalylglycine. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Strain-Damerell, C, Gileadi, C, Szykowska, A, Kupinska, K, Kopec, J, Krojer, T, Steuber, H, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5A3T
 
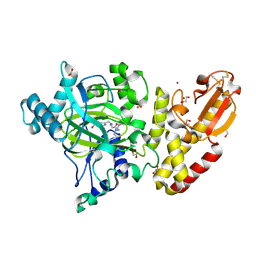 | | Crystal structure of human PLU-1 (JARID1B) in complex with KDM5-C49 (2-(((2-((2-(dimethylamino)ethyl)(ethyl)amino)-2-oxoethyl)amino)methyl) isonicotinic acid). | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, BurgessBrown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-17 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5A3P
 
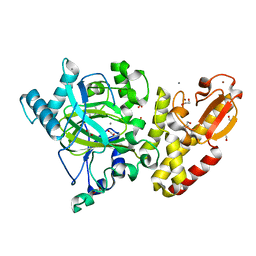 | | Crystal structure of the catalytic domain of human PLU1 (JARID1B). | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Tallant, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5A3W
 
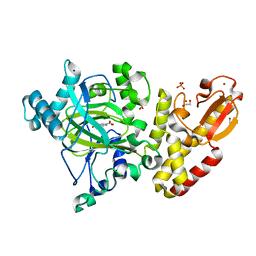 | | Crystal structure of human PLU-1 (JARID1B) in complex with Pyridine-2, 6-dicarboxylic Acid (PDCA) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FPU
 
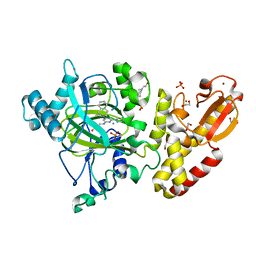 | | Crystal structure of human JARID1B in complex with GSKJ1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Nowak, R, Johansson, C, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5A3N
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with KDOAM25a | | Descriptor: | 1,2-ETHANEDIOL, 2-[[[2-[2-(dimethylamino)ethyl-ethyl-amino]-2-oxidanylidene-ethyl]amino]methyl]pyridine-4-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Nuzzi, A, Ruda, G.F, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-08 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells.
Cell Chem Biol, 24, 2017
|
|
5FPV
 
 | | Crystal structure of human JMJD2A in complex with compound KDOAM20A | | Descriptor: | 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, MANGANESE (II) ION, ... | | Authors: | Srikannathasan, V, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FUP
 
 | | Crystal structure of human JARID1B in complex with 2-oxoglutarate. | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Talon, R, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FV3
 
 | | Crystal structure of human JARID1B construct c2 in complex with N- Oxalylglycine. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Talon, R, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FUN
 
 | | Crystal structure of human JARID1B in complex with GSK467 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(1-benzyl-1H-pyrazol-4-yl)oxy]pyrido[3,4-d]pyrimidin-4(3H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Oppermann, U. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
