5Z1X
 
 | | Crystal Structure of Laccase from Cerrena sp. RSD1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Lee, C.C, Wu, M.H, Ho, T.H, Wang, A.H.J. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kinetic analysis and structural studies of a high-efficiency laccase fromCerrenasp. RSD1.
FEBS Open Bio, 8, 2018
|
|
5Z22
 
 | | Crystal Structure of Laccase from Cerrena sp. RSD1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Lee, C.C, Wu, M.H, Ho, T.H, Wang, A.H.J. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhancement of laccase activity by pre-incubation with organic solvents.
Sci Rep, 9, 2019
|
|
3UQH
 
 | | Crystal structure of aba receptor pyl10 (apo) | | Descriptor: | Abscisic acid receptor PYL10, SULFATE ION | | Authors: | Sun, D.M, Wang, H.P, Wu, M.H, Zang, J.Y, Tian, C.L. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Aba Receptor Pyl10
To be Published
|
|
3R6P
 
 | | Crystal structure of abscisic acid-bound PYL10 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL10 | | Authors: | Sun, D.M, Wu, M.H, Wang, H.P, Zang, J.Y, Tian, C.L. | | Deposit date: | 2011-03-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of abscisic acid-bound PYL10
To be Published
|
|
4WGI
 
 | | A Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1) | | Descriptor: | (2S)-2-[(2S,3R)-10-{[(4-fluorophenyl)sulfonyl]amino}-3-methyl-2-[(methyl{[4-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-5(6H)-yl]propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Fairman, J.W, Fang, C, D'Souza, B, Fulroth, B, Leed, A, McCarren, P, Wang, L, Wang, Y, Kaushik, V, Palmer, M, Wei, G, Golub, T.R, Hubbard, B.K, Serrano-Wu, M.H. | | Deposit date: | 2014-09-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1).
Acs Med.Chem.Lett., 5, 2014
|
|
8XBF
 
 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O5C2, heavy chain, ... | | Authors: | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
5EK3
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-[(5~{S})-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5EK2
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | 1-cyclohexyl-2-[(5~{S})-6-fluoranyl-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanone, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5EK4
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-[(5~{S})-6-fluoranyl-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, S.Y, Peng, Y.H, Wu, J.S. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5ETW
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-pyrido[3,4-b]indol-9-yl-ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, S.Y, Peng, Y.H, Wu, J.S. | | Deposit date: | 2015-11-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1.
J.Med.Chem., 59, 2016
|
|
6KLA
 
 | | Crystal structure of human c-KIT kinase domain in complex with compound 15a | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-[6-(4-ethylpiperazin-1-yl)-2-methyl-pyrimidin-4-yl]-5-pyridin-4-yl-1,3-thiazol-2-amine | | Authors: | Wu, T.S, Peng, Y.H, Hsueh, C.C, Wu, S.Y. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Identification of a Multitargeted Tyrosine Kinase Inhibitor for the Treatment of Gastrointestinal Stromal Tumors and Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
6KOF
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 47 | | Descriptor: | 1-(4-cyanophenyl)-3-[[3-(2-cyclopropylethynyl)imidazo[2,1-b][1,3]thiazol-5-yl]methyl]thiourea, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-08-09 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6KW7
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 12 | | Descriptor: | 3-(4-bromophenyl)imidazo[2,1-b][1,3]thiazole, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-09-06 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6KPS
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 36 | | Descriptor: | 1-(4-cyanophenyl)-3-[[3-(2-cyclopropylethynyl)imidazo[2,1-b][1,3]thiazol-5-yl]methyl]urea, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-08-16 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
5BOE
 
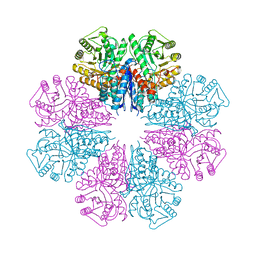 | | Crystal structure of Staphylococcus aureus enolase in complex with PEP | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, C.L, Wu, Y.F, Han, L, Wu, M.H, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5BOF
 
 | | Crystal Structure of Staphylococcus aureus Enolase | | Descriptor: | Enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Wu, Y.F, Wang, C.L, Wu, M.H, Han, L, Zhang, X, Zang, J.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Octameric structure of Staphylococcus aureus enolase in complex with phosphoenolpyruvate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WMW
 
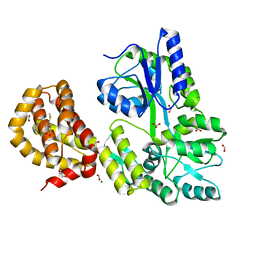 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMS
 
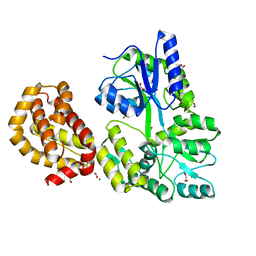 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMR
 
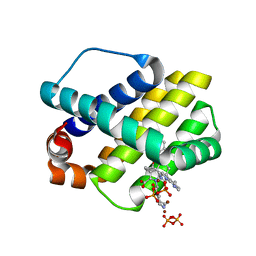 | | STRUCTURE OF MCL1 BOUND TO BRD inhibitor ligand 1 AT 1.7A | | Descriptor: | 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, PYROPHOSPHATE 2-, ... | | Authors: | CLIFTON, M.C, EDWARDS, T.E. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMX
 
 | | The structure of MBP-MCL1 bound to ligand 6 at 2.0A | | Descriptor: | 4-ethenyl-2-[(phenylsulfonyl)amino]benzoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMT
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 1 AT 2.35A | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMV
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 4 AT 2.4A | | Descriptor: | 3-chloro-6-fluoro-1-benzothiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Moulin, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMU
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 2 AT 1.55A | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Faiman, J.W. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
7E38
 
 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with yatein and succinate | | Descriptor: | (3~{R},4~{R})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, (3~{S},4~{S})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, Deoxypodophyllotoxin synthase, ... | | Authors: | Wu, M.-H, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E37
 
 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Deoxypodophyllotoxin synthase, FE (III) ION | | Authors: | Wu, M.-H, Lin, H.-Y, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
