2ARA
 
 | |
2ARC
 
 | |
4DHJ
 
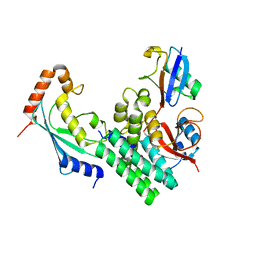 | | The structure of a ceOTUB1 ubiquitin aldehyde UBC13~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
4DHZ
 
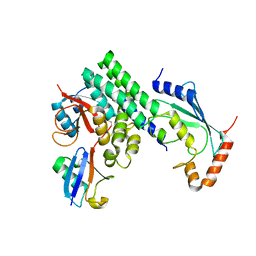 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBC13~Ub | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
4FH1
 
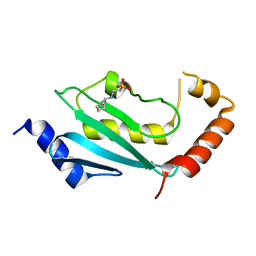 | | S. cerevisiae Ubc13-N79A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Ubiquitin-conjugating enzyme E2 13 | | Authors: | Berndsen, C.E, Wolberger, C, Wiener, R. | | Deposit date: | 2012-06-05 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A conserved asparagine has a structural role in ubiquitin-conjugating enzymes.
Nat.Chem.Biol., 9, 2013
|
|
4FIP
 
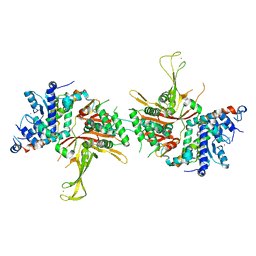 | | Structure of the SAGA Ubp8(S144N)/Sgf11(1-72, Delta-ZnF)/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
4FK5
 
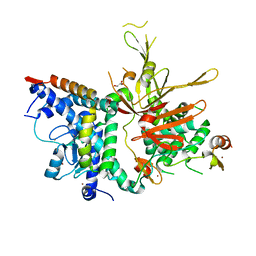 | | Structure of the SAGA Ubp8(S144N)/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
4FJC
 
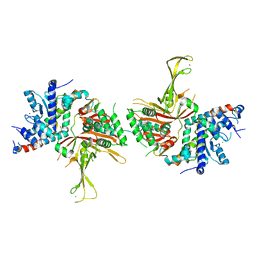 | | Structure of the SAGA Ubp8/Sgf11(1-72, Delta-ZnF)/Sus1/Sgf73 DUB module | | Descriptor: | GLYCEROL, Protein SUS1, SAGA-associated factor 11, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-11 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.826 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
1K7A
 
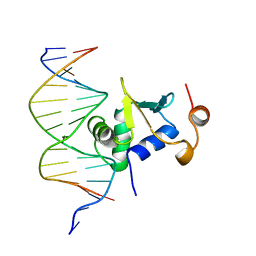 | | Ets-1(331-440)+GGAG duplex | | Descriptor: | C-ets-1 Protein, DNA (5'-D(*CP*AP*CP*AP*TP*CP*TP*CP*CP*GP*GP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*CP*CP*GP*GP*AP*GP*AP*TP*GP*T)-3') | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1K61
 
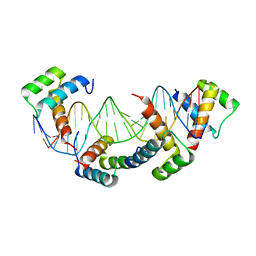 | | MATALPHA2 HOMEODOMAIN BOUND TO DNA | | Descriptor: | 5'-D(*(5IU)P*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*AP*CP*AP*TP*G)-3', 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*CP*AP*CP*GP*C)-3', Mating-type protein alpha-2 | | Authors: | Aishima, J, Gitti, R.K, Noah, J.E, Gan, H.H, Schlick, T, Wolberger, C. | | Deposit date: | 2001-10-14 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Hoogsteen base pair embedded in undistorted B-DNA
NUCLEIC ACIDS RES., 30, 2002
|
|
1K78
 
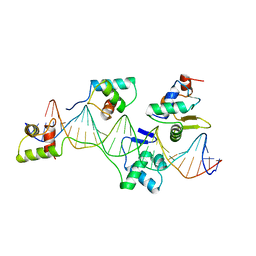 | | Pax5(1-149)+Ets-1(331-440)+DNA | | Descriptor: | C-ets-1 Protein, Paired Box Protein Pax5, Pax5/Ets Binding Site on the mb-1 promoter | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1K79
 
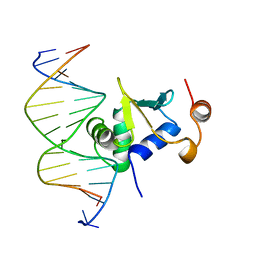 | | Ets-1(331-440)+GGAA duplex | | Descriptor: | C-ets-1 protein, DNA (5'-D(*CP*AP*CP*AP*TP*TP*TP*CP*CP*GP*GP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*CP*CP*GP*GP*AP*AP*AP*TP*GP*T)-3') | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1LE8
 
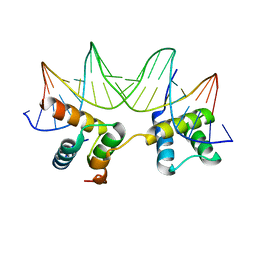 | | Crystal Structure of the MATa1/MATalpha2-3A Heterodimer Bound to DNA Complex | | Descriptor: | 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*AP*AP*AP*TP*TP*TP*AP*CP*AP*TP*CP*A)-3', 5'-D(*TP*TP*GP*AP*TP*GP*TP*AP*AP*AP*TP*TP*TP*TP*TP*AP*CP*AP*TP*G)-3', MATING-TYPE PROTEIN A-1, ... | | Authors: | Ke, A, Mathias, J.R, Vershon, A.K, Wolberger, C. | | Deposit date: | 2002-04-09 | | Release date: | 2002-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Characterization of the DNA Binding Properties of a Triple Alanine Mutant of MATalpha2
Structure, 10, 2002
|
|
1MH3
 
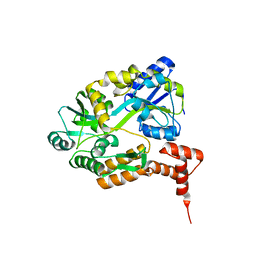 | | maltose binding-a1 homeodomain protein chimera, crystal form I | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
1MH4
 
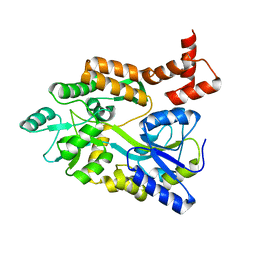 | | maltose binding-a1 homeodomain protein chimera, crystal form II | | Descriptor: | maltose binding-a1 homeodomain protein chimera | | Authors: | Ke, A, Wolberger, C. | | Deposit date: | 2002-08-19 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into binding cooperativity of MATa1/MATalpha2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera
Protein Sci., 12, 2003
|
|
1MDM
 
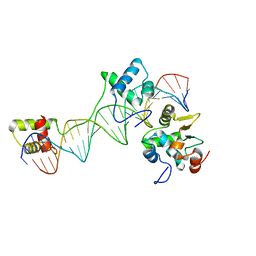 | | INHIBITED FRAGMENT OF ETS-1 AND PAIRED DOMAIN OF PAX5 BOUND TO DNA | | Descriptor: | C-ETS-1 PROTEIN, PAIRED BOX PROTEIN PAX-5, PAX5/ETS BINDING SITE ON THE MB-1 PROMOTER | | Authors: | Garvie, C.W, Pufall, M.A, Graves, B.J, Wolberger, C. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STRUCTURAL ANALYSIS OF THE AUTOINHIBITION OF ETS-1 AND ITS ROLE IN PROTEIN PARTNERSHIPS
J.Biol.Chem., 277, 2002
|
|
1MD0
 
 | |
1MA3
 
 | | Structure of a Sir2 enzyme bound to an acetylated p53 peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cellular tumor antigen p53, Transcriptional regulatory protein, ... | | Authors: | Avalos, J.L, Celic, I, Muhammad, S, Cosgrove, M.S, Boeke, J.D, Wolberger, C. | | Deposit date: | 2002-07-31 | | Release date: | 2002-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Sir2 enzyme bound to an acetylated p53 peptide
Mol.Cell, 10, 2002
|
|
2CRO
 
 | |
2H3B
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor 1 | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H2D
 
 | |
2H2F
 
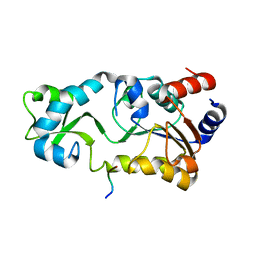 | |
2H4J
 
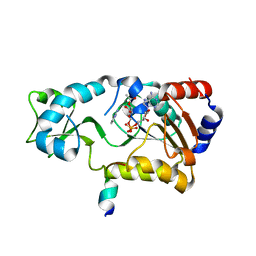 | | Sir2-deacetylated peptide (from enzymatic turnover in crystal) | | Descriptor: | 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, Cellular tumor antigen p53, NAD-dependent deacetylase, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
2H59
 
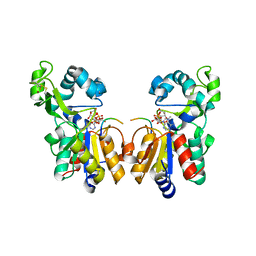 | | Sir2 H116A-deacetylated p53 peptide-3'-o-acetyl ADP ribose | | Descriptor: | (2S,3S,4R,5S)-2-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-4,5-DIHYDROXYTETRAHYDROFURAN-3-YL ACETATE, ADENOSINE-5-DIPHOSPHORIBOSE, Cellular tumor antigen p53, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-25 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
