1IHR
 
 | | Crystal structure of the dimeric C-terminal domain of TonB | | Descriptor: | BROMIDE ION, TonB protein | | Authors: | Chang, C, Mooser, A, Pluckthun, A, Wlodawer, A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold.
J.Biol.Chem., 276, 2001
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1FSI
 
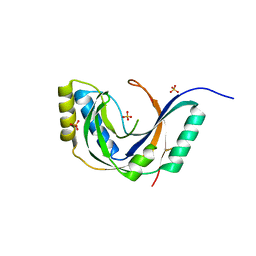 | | CRYSTAL STRUCTURE OF CYCLIC NUCLEOTIDE PHOSPHODIESTERASE OF APPR>P FROM ARABIDOPSIS THALIANA | | Descriptor: | CYCLIC PHOSPHODIESTERASE, SULFATE ION | | Authors: | Hofmann, A, Zdanov, A, Genschik, P, Filipowicz, W, Ruvinov, S, Wlodawer, A. | | Deposit date: | 2000-09-10 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction.
EMBO J., 19, 2000
|
|
2CRK
 
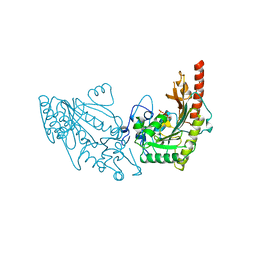 | | MUSCLE CREATINE KINASE | | Descriptor: | PROTEIN (CREATINE KINASE), SULFATE ION | | Authors: | Rao, J.K, Bujacz, G, Wlodawer, A. | | Deposit date: | 1998-09-28 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of rabbit muscle creatine kinase.
FEBS Lett., 439, 1998
|
|
1RRE
 
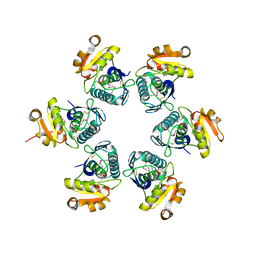 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1RR9
 
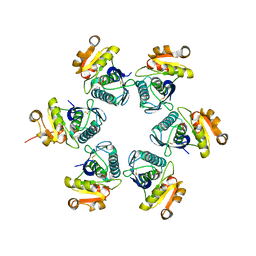 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1LVM
 
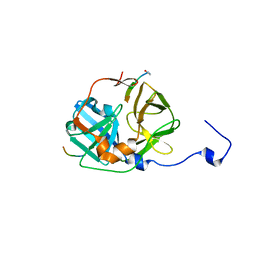 | | CATALYTICALLY ACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH PRODUCT | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
1LVB
 
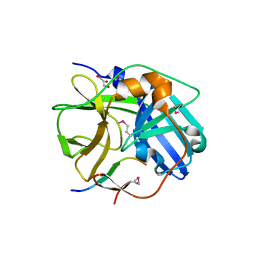 | | CATALYTICALLY INACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH SUBSTRATE | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), GLYCEROL, OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
1HO3
 
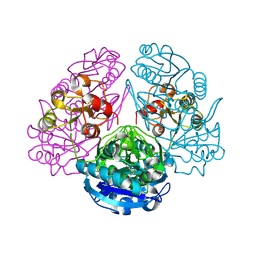 | | CRYSTAL STRUCTURE ANALYSIS OF E. COLI L-ASPARAGINASE II (Y25F MUTANT) | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE II | | Authors: | Jaskolski, M, Kozak, M, Lubkowski, P, Palm, J.G, Wlodawer, A. | | Deposit date: | 2000-12-08 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two highly homologous bacterial L-asparaginases: a case of enantiomorphic space groups.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1YG9
 
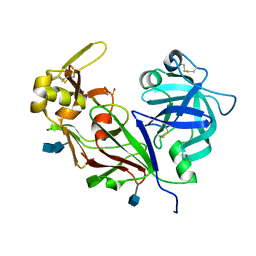 | | The structure of mutant (N93Q) of bla g 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aspartic protease Bla g 2, ... | | Authors: | Li, M, Gustchina, A, Wuenschmann, S, Pomes, A, Wlodawer, A. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition.
J.Mol.Biol., 348, 2005
|
|
1N1F
 
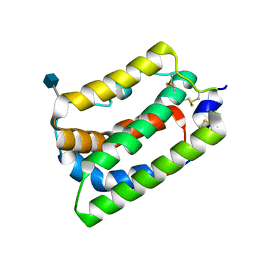 | | Crystal Structure of Human Interleukin-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, interleukin-19 | | Authors: | Chang, C, Magracheva, E, Kozlov, S, Fong, S, Tobin, G, Kotenko, S, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-10-17 | | Release date: | 2003-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of interleukin-19 defines a new subfamily of helical cytokines
J.Biol.Chem., 278, 2003
|
|
1ILK
 
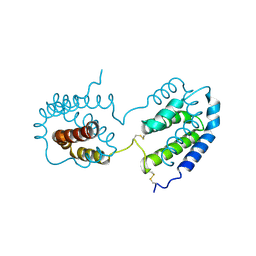 | |
1TQM
 
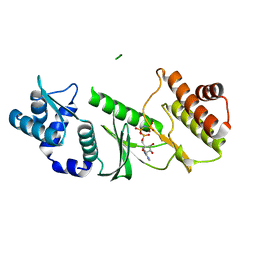 | |
1TQI
 
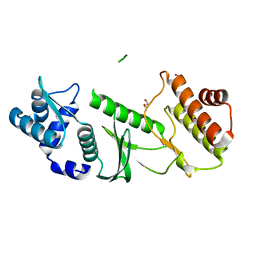 | |
1TQP
 
 | |
1TOL
 
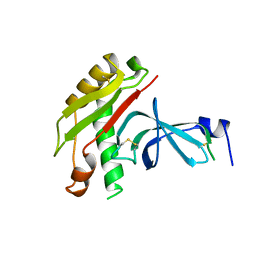 | | FUSION OF N-TERMINAL DOMAIN OF THE MINOR COAT PROTEIN FROM GENE III IN PHAGE M13, AND C-TERMINAL DOMAIN OF E. COLI PROTEIN-TOLA | | Descriptor: | PROTEIN (FUSION PROTEIN CONSISTING OF MINOR COAT PROTEIN, GLYCINE RICH LINKER, TOLA, ... | | Authors: | Lubkowski, J, Wlodawer, A, Hennecke, F, Plueckthun, A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA.
Structure Fold.Des., 7, 1999
|
|
1ODX
 
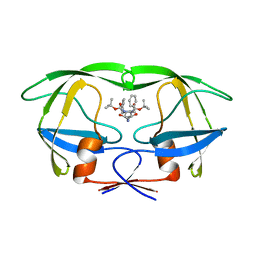 | | HIV-1 Proteinase mutant A71T, V82A | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Kervinen, J, Thanki, N, Zdanov, A, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
2GTY
 
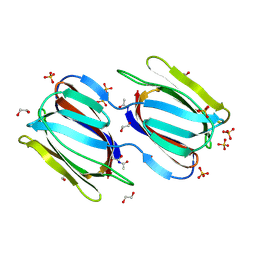 | | Crystal structure of unliganded griffithsin | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, SULFATE ION | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
2GUC
 
 | |
2GUD
 
 | |
2GUX
 
 | | Selenomethionine derivative of griffithsin | | Descriptor: | SULFATE ION, griffithsin | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-05-01 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
2GUE
 
 | |
2HEX
 
 | |
1G3P
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAINS OF BACTERIOPHAGE MINOR COAT PROTEIN G3P | | Descriptor: | MINOR COAT PROTEIN, SULFATE ION | | Authors: | Lubkowski, J, Hennecke, F, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1997-12-22 | | Release date: | 1998-01-28 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p.
Nat.Struct.Biol., 5, 1998
|
|
1ODW
 
 | | Native HIV-1 Proteinase | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Thanki, N, Kervinen, J, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
