4E5L
 
 | | Crystal structure of avian influenza virus PAn bound to compound 6 | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5J
 
 | | Crystal structure of avian influenza virus PAn bound to compound 5 | | Descriptor: | 2-[3-(acetylamino)phenyl]-5-hydroxy-6-oxo-1,6-dihydropyrimidine-4-carboxylic acid, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5I
 
 | | Crystal structure of avian influenza virus PAn bound to compound 4 | | Descriptor: | 5-hydroxy-2-(1-methyl-1H-imidazol-4-yl)-6-oxo-1,6-dihydropyrimidine-4-carboxylic acid, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
6V6X
 
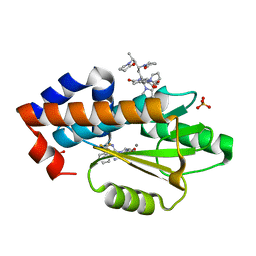 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000988632 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VG9
 
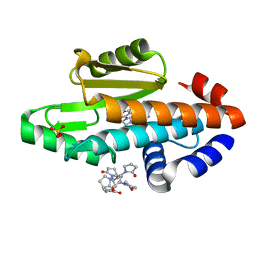 | | The crystal structure of the 2009 H1N1/California PA endonuclease I38T mutant in complex with SJ000986248 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6V9E
 
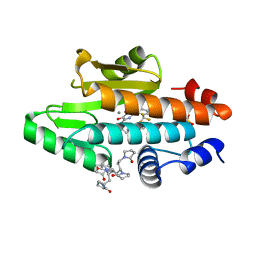 | | The crystal structure of the 2009 H1N1 PA endonuclease wild type in complex with SJ000988632 | | Descriptor: | 1,1',1'',1''',1''''-[(3R,5S,7S,9R)-decane-1,3,5,7,9-pentayl]penta(pyrrolidin-2-one), MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-13 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VBR
 
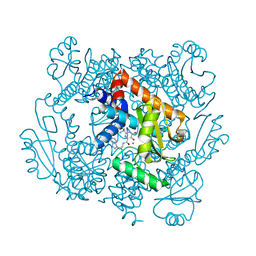 | | The crystal structure of the 2009 H1N1/California PA endonuclease wild type in complex with SJ000986248 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-19 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
2B4Q
 
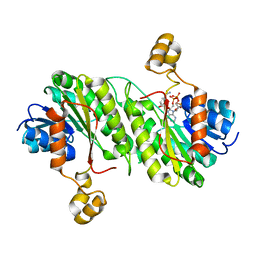 | | Pseudomonas aeruginosa RhlG/NADP active-site complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Rhamnolipids biosynthesis 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Miller, D.J, White, S.W. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of RhlG, an Essential beta-Ketoacyl Reductase in the Rhamnolipid Biosynthetic Pathway of Pseudomonas aeruginosa.
J.Biol.Chem., 281, 2006
|
|
6W7A
 
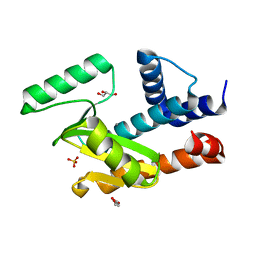 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant E119D bound to DNA oligomer TAGCAT (uncleaved, 5mM overnight DNA soak) | | Descriptor: | DNA (5'-D(P*TP*AP*GP*CP*AP*T)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
6VL3
 
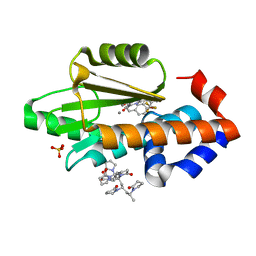 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986436 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-22 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VJH
 
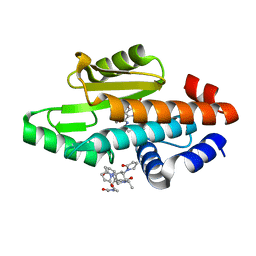 | | The crystal structure of the 2009 H1N1/California PA endonuclease wild type in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VIV
 
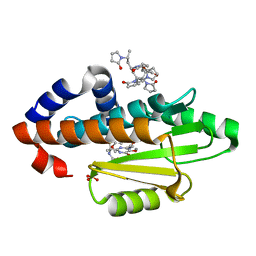 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000986192 | | Descriptor: | 2-[2-[(cyclohexylmethyl-$l^{3}-oxidanyl)carbonylamino]propan-2-yl]-~{N}-[2-(5-methoxy-4-oxidanyl-cyclohexa-1,3,5-trien-1-yl)ethyl]-5-oxidanyl-6-oxidanylidene-pyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-14 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6W6B
 
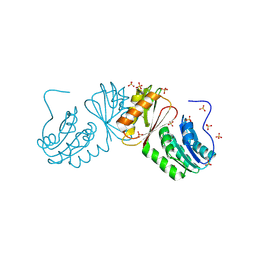 | | The X-ray crystal structure of the C-terminus domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 328-548) protein to 1.40 Angstrom resolution | | Descriptor: | SULFATE ION, SaFakA-Cterminus domain | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The X-ray crystal structure of the C-terminus domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 328-548) protein to 1.40 Angstrom resolution
To Be Published
|
|
2ALM
 
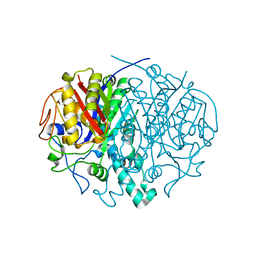 | | Crystal structure analysis of a mutant beta-ketoacyl-[acyl carrier protein] synthase II from Streptococcus pneumoniae | | Descriptor: | 3-oxoacyl-(acyl-carrier-protein) synthase II, MAGNESIUM ION | | Authors: | Zhang, Y.M, Hurlbert, J, White, S.W, Rock, C.O. | | Deposit date: | 2005-08-07 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of the active site water, histidine 303, and phenylalanine 396 in the catalytic mechanism of the elongation condensing enzyme of Streptococcus pneumoniae.
J.Biol.Chem., 281, 2006
|
|
6WHM
 
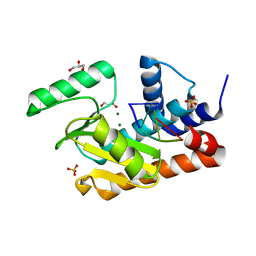 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomer TAGC (cleaved TTAGCATT, 5mM overnight DNA soak) | | Descriptor: | DNA (5'-D(P*TP*AP*GP*C)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
4HNJ
 
 | |
6WS3
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomers TG and AGCA (from cleaved GTGAGCAGTG) | | Descriptor: | DNA (5'-D(P*TP*G)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
6WIJ
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant I38T in complex with SJ000986448 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-04-10 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6WJ4
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type in complex with SJ000986448 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
4JUM
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, clade 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Coop, T, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2013-03-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | To be published
To be Published
|
|
4JUL
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, clade 2.3.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Coop, T, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2013-03-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7927 Å) | | Cite: | To be published
To be Published
|
|
4JUK
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, clade 2.3.2.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Coop, T, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2013-03-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7502 Å) | | Cite: | To be published
To be Published
|
|
4JUN
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, clade 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Coop, T, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2013-03-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3398 Å) | | Cite: | To be published
To be Published
|
|
2OCA
 
 | | The crystal structure of T4 UvsW | | Descriptor: | ATP-dependent DNA helicase uvsW | | Authors: | Kerr, I.D, White, S.W. | | Deposit date: | 2006-12-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Analyses of UvsW and UvsW.1 from Bacteriophage T4.
J.Biol.Chem., 282, 2007
|
|
5V85
 
 | | The crystal structure of the protein of DegV family COG1307 from Ruminococcus gnavus ATCC 29149 (alternative refinement of PDB 3JR7 with Vaccenic acid) | | Descriptor: | EDD domain protein, DegV family, PHOSPHATE ION, ... | | Authors: | Cuypers, M.G, Ericson, M, subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-03-21 | | Release date: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
