2MJ2
 
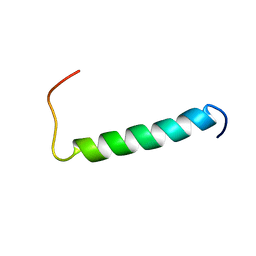 | | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, S.A, Abou-Gharbia, M, Childers, W, White, M, Bouaziz, S, Safak, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix
J.Virol., 2014
|
|
1MM8
 
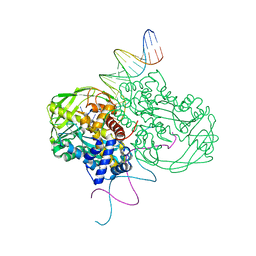 | | Crystal structure of Tn5 Transposase complexed with ME DNA | | Descriptor: | MANGANESE (II) ION, ME DNA non-transferred strand, ME DNA transferred strand, ... | | Authors: | Steiniger-White, M, Bhasin, A, Lovell, S, Rayment, I, Reznikoff, W.S. | | Deposit date: | 2002-09-03 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for "unseen" Transposase--DNA contacts
J.Mol.Biol., 322, 2002
|
|
1MUS
 
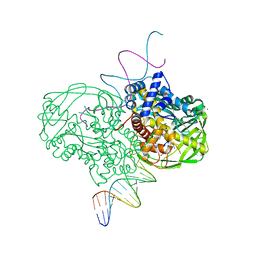 | | crystal structure of Tn5 transposase complexed with resolved outside end DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA non-transferred strand, DNA transferred strand, ... | | Authors: | Holden, H.M, Thoden, J.B, Steiniger-White, M, Reznikoff, W.S, Lovell, S, Rayment, I. | | Deposit date: | 2002-09-24 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure/function insights into Tn5 transposition.
Curr.Opin.Struct.Biol., 14, 2004
|
|
1R8T
 
 | | Solution structures of high affinity miniprotein ligands to Streptavidin | | Descriptor: | MP1 | | Authors: | Luo, J, Mukherjee, M, Fan, X, Yang, H, Liu, D, Khan, R, White, M, Fox, R.O. | | Deposit date: | 2003-10-28 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure-based design of high affinity miniprotein ligands
To be Published
|
|
2C0J
 
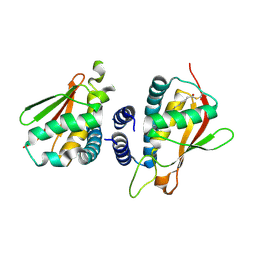 | | Crystal structure of the bet3-trs33 heterodimer | | Descriptor: | PALMITIC ACID, R32611_2, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3 | | Authors: | Kim, M.-S, Yi, M.-J, Lee, K.-H, Wagner, J, Munger, C, Kim, Y.-G, Whiteway, M, Cygler, M, Oh, B.-H, Sacher, M. | | Deposit date: | 2005-09-03 | | Release date: | 2006-02-07 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies Reveal a Specific Interaction between Trapp Subunits Trs33P and Bet3P
Traffic, 6, 2005
|
|
1X9X
 
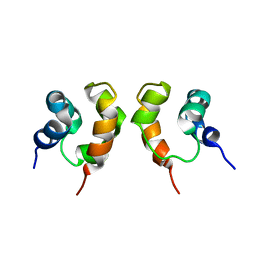 | | Solution Structure of Dimeric SAM Domain from MAPKKK Ste11 | | Descriptor: | Serine/threonine-protein kinase STE11 | | Authors: | Bhattacharjya, S, Xu, P, Gingras, R, Shaykhutdinov, R, Wu, C, Whiteway, M, Ni, F. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: implications for Ste11 activation and signal transmission through the Ste50-Ste11 complex.
J.Mol.Biol., 344, 2004
|
|
6VG5
 
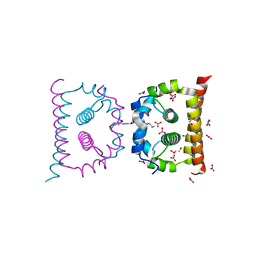 | | DengueV-2 Capsid ST148 inhibitor Complex | | Descriptor: | 3-amino-N-(5-phenyl-1,3,4-thiadiazol-2-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]thieno[3,2-e]pyridine-2-carboxamide, Capsid premembrane protein, GLYCEROL, ... | | Authors: | White, M, Xia, H, Shi, P. | | Deposit date: | 2020-01-07 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A cocrystal structure of dengue capsid protein in complex of inhibitor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VSO
 
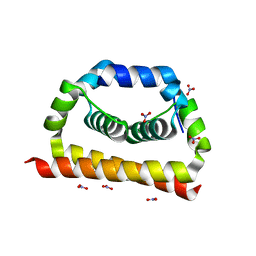 | | DengueV-2 Capsid Structure | | Descriptor: | Capsid premembrane protein, GLYCEROL, NITRATE ION | | Authors: | White, M, Xia, H, Shi, P. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A cocrystal structure of dengue capsid protein in complex of inhibitor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5M1B
 
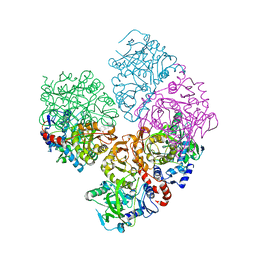 | | Crystal structure of C-terminally tagged apo-UbiD from E. coli | | Descriptor: | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase | | Authors: | White, M, Leys, D. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
5TZB
 
 | | Burkholderia sp. beta-aminopeptidase | | Descriptor: | CALCIUM ION, D-aminopeptidase | | Authors: | McGowan, S, Drinkwater, N, John, M, Dumsday, G. | | Deposit date: | 2016-11-21 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Crystal structure of a beta-aminopeptidase from an Australian Burkholderia sp.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1E59
 
 | |
8V1L
 
 | |
8EUG
 
 | | Ytm1 associated nascent 60S ribosome State 3 | | Descriptor: | 60S ribosomal protein L10-A, 60S ribosomal protein L11-A, 60S ribosomal protein L13, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETJ
 
 | | Fkbp39 associated 60S nascent ribosome State 2 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESQ
 
 | | Ytm1 associated nascent 60S ribosome State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETC
 
 | | Fkbp39 associated nascent 60S ribosome State 4 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUY
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUI
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 3 | | Descriptor: | 60S ribosomal protein L10-A, 60S ribosomal protein L11-A, 60S ribosomal protein L13, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETH
 
 | | Ytm1 associated 60S nascent ribosome State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESR
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETI
 
 | | Fkbp39 associated 60S nascent ribosome State 1 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUP
 
 | | Ytm1 associated 60S nascent ribosome State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETG
 
 | | Fkbp39 associated 60S nascent ribosome State 3 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EV3
 
 | | Ytm1 associated 60S nascent ribosome (-Fkbp39) State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
2XVO
 
 | | SSO1725, a protein involved in the CRISPR/Cas pathway | | Descriptor: | BETA-MERCAPTOETHANOL, SSO1725, SULFATE ION | | Authors: | Reeks, J, Liu, H, Naismith, J, White, M, McMahon, S. | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-29 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and Mechanism of the Cmr Complex for Crispr-Mediated Antiviral Immunity.
Mol.Cell, 45, 2012
|
|
