6MFL
 
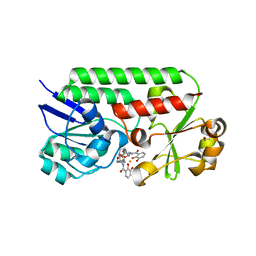 | | Structure of siderophore binding protein BauB bound to a complex between two molecules of acinetobactin and ferric iron. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Siderophore Binding Protein BauB, ... | | Authors: | Gulick, A.M, Bailey, D.C, Wencewicz, T.A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Siderophore Binding Protein BauB Bound to an Unusual 2:1 Complex Between Acinetobactin and Ferric Iron.
Biochemistry, 57, 2018
|
|
6N8E
 
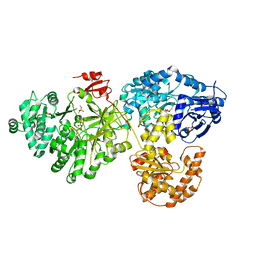 | | Crystal structure of holo-ObiF1, a five domain nonribosomal peptide synthetase from Burkholderia diffusa | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 4-(4-nitrophenyl)-L-threonine, CHLORIDE ION, ... | | Authors: | Kreitler, D.F, Wencewicz, T.A, Gulick, A.M. | | Deposit date: | 2018-11-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of N-acyl-alpha-amino-beta-lactone formation catalyzed by a nonribosomal peptide synthetase.
Nat Commun, 10, 2019
|
|
8TWF
 
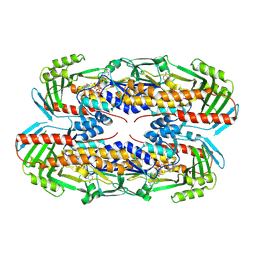 | |
8TWG
 
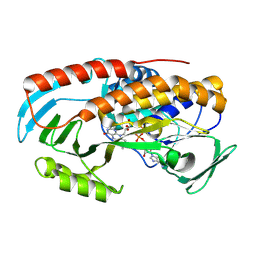 | |
6WG9
 
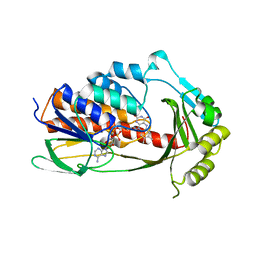 | | Crystal structure of tetracycline destructase Tet(X7) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Tetracycline destructase Tet(X7) | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance.
Commun Biol, 3, 2020
|
|
8ER1
 
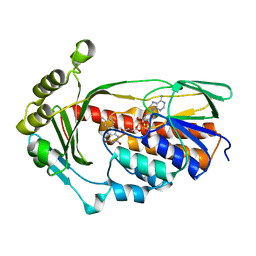 | | X-ray crystal structure of Tet(X6) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
8ER0
 
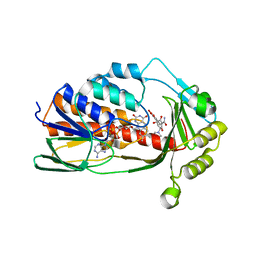 | | X-ray crystal structure of Tet(X6) bound to anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2022-10-11 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.
Commun Biol, 6, 2023
|
|
7LRN
 
 | |
7TGL
 
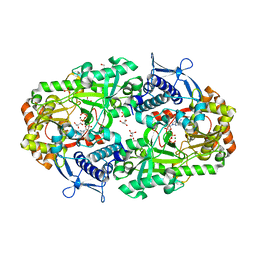 | |
7TGK
 
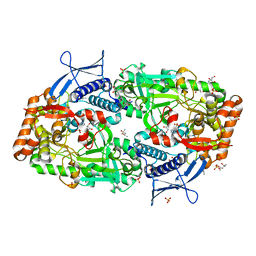 | | Crystal structure of ATP bound DesD, the desferrioxamine synthetase from the Streptomyces griseoflavus ferrimycin biosynthetic pathway | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2022-01-07 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
7TGN
 
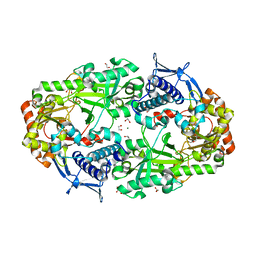 | | Crystal structure of DesD, the desferrioxamine synthetase from the Streptomyces violaceus salmycin biosynthetic pathway | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Desferrioxamine synthetase DesD, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2022-01-07 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
7TGM
 
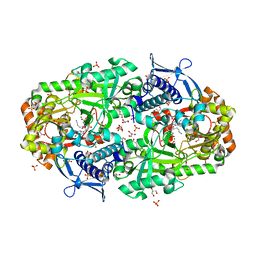 | |
7TGJ
 
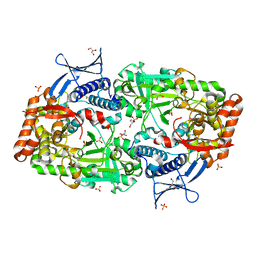 | |
5TUM
 
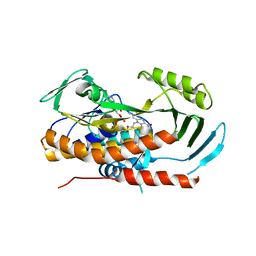 | | Crystal structure of tetracycline destructase Tet(56) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tetracycline destructase Tet(56) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUF
 
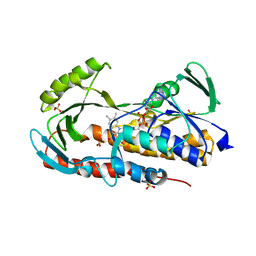 | | Crystal structure of tetracycline destructase Tet(50) in complex with anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUL
 
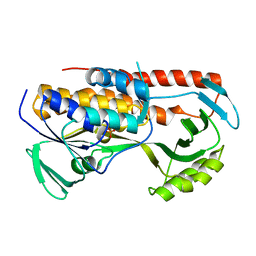 | |
5TUK
 
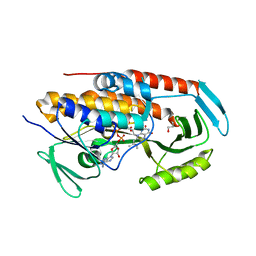 | | Crystal structure of tetracycline destructase Tet(51) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Tetracycline destructase Tet(51) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUI
 
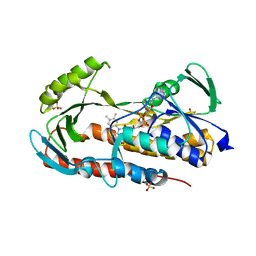 | | Crystal structure of tetracycline destructase Tet(50) in complex with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUE
 
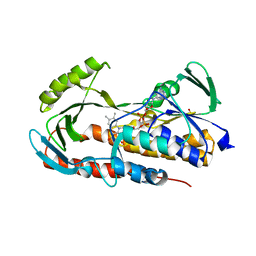 | | Crystal structure of tetracycline destructase Tet(50) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Tetracycline destructase Tet(50) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
