1BHS
 
 | | HUMAN ESTROGENIC 17BETA-HYDROXYSTEROID DEHYDROGENASE | | Descriptor: | 17BETA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Ghosh, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human estrogenic 17 beta-hydroxysteroid dehydrogenase at 2.20 A resolution.
Structure, 3, 1995
|
|
1CLE
 
 | | STRUCTURE OF UNCOMPLEXED AND LINOLEATE-BOUND CANDIDA CYLINDRACEA CHOLESTEROL ESTERASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL ESTERASE, ... | | Authors: | Ghosh, D. | | Deposit date: | 1995-02-08 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of uncomplexed and linoleate-bound Candida cylindracea cholesterol esterase.
Structure, 3, 1995
|
|
8DL1
 
 | | BoGH13ASus-E523Q from Bacteroides ovatus bound to maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha amylase, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-07-06 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
8DL2
 
 | | BoGH13ASus from Bacteroides ovatus bound to acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-07-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
8DGE
 
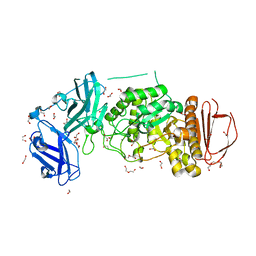 | | BoGH13ASus from Bacteroides ovatus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Alpha amylase, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-06-23 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
4W8K
 
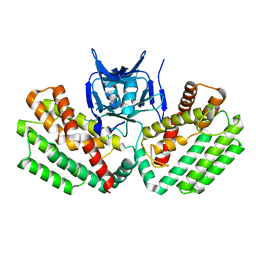 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
2FIH
 
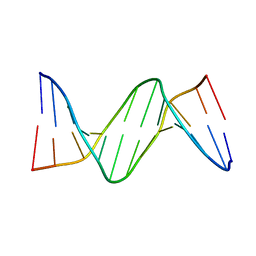 | |
2FII
 
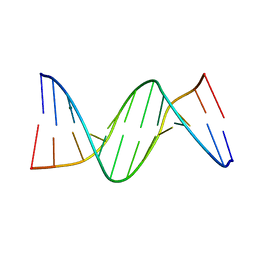 | |
2FIJ
 
 | |
2FIL
 
 | |
6V0A
 
 | | Crystal structure of cytochrome c nitrite reductase from the bacterium Geobacter lovleyi with bound sulfate | | Descriptor: | HEME C, Nitrite reductase (cytochrome; ammonia-forming), SULFATE ION | | Authors: | Satyanarayana, L, Campecino, J, Hegg, L.H, Hu, J. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cytochromecnitrite reductase from the bacteriumGeobacter lovleyirepresents a new NrfA subclass.
J.Biol.Chem., 295, 2020
|
|
2HSD
 
 | |
5KZH
 
 | |
5L2F
 
 | | High Resolution Structure of Acinetobacter baumannii beta-lactamase OXA-51 I129L/K83D bound to doripenem | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ACETATE ION, Beta-lactamase, ... | | Authors: | June, C.M, Powers, R.A, Leonard, D.A, Muckenthaler, T. | | Deposit date: | 2016-08-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structure of a doripenem-bound OXA-51 class D beta-lactamase variant with enhanced carbapenemase activity.
Protein Sci., 25, 2016
|
|
6WN5
 
 | | 1.52 Angstrom Resolution Crystal Structure of Transcriptional Regulator HdfR from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Transcriptional regulator HdfR | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6WN8
 
 | | 2.70 Angstrom Resolution Crystal Structure of Uracil Phosphoribosyl Transferase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, SULFATE ION, Uracil phosphoribosyltransferase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6X1L
 
 | | The crystal structure of a functional uncharacterized protein KP1_0663 from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | Descriptor: | WbbZ protein | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6X3K
 
 | |
6X3Q
 
 | |
7KMJ
 
 | | Hsa Siglec and Unique domains in complex with Sialyl Lewis C | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Stubbs, H.E, Iverson, T.M. | | Deposit date: | 2020-11-03 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
7RJJ
 
 | | Crystal Structure of the Peptidoglycan Binding Domain of the Outer Membrane Protein (OmpA) from Klebsiella pneumoniae with bound D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, OmpA family protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7RPY
 
 | | X25-2 domain of Sca5 from Ruminococcus bromii | | Descriptor: | ACETATE ION, Cohesin-containing protein, GLYCEROL, ... | | Authors: | Cerqueira, F, Koropatkin, N. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome.
J.Biol.Chem., 298, 2022
|
|
7RAW
 
 | |
7RFT
 
 | |
7TA0
 
 | | Human Ornithine Aminotransferase (hOAT) soaked with 5-aminovaleric acid | | Descriptor: | 5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Liu, D. | | Deposit date: | 2021-12-20 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Determination of the pH dependence, substrate specificity, and turnovers of alternative substrates for human ornithine aminotransferase.
J.Biol.Chem., 298, 2022
|
|
