4MWA
 
 | | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Brunzelle, J.S, Xu, X, Cui, H, Maltseva, N, Bishop, B, Kwon, K, Savchenko, A, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
3N0L
 
 | | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni
TO BE PUBLISHED
|
|
3OET
 
 | | D-Erythronate-4-Phosphate Dehydrogenase complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Edwards, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-13 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | D-Erythronate-4-Phosphate Dehydrogenase complexed with NAD
To be Published
|
|
4MLG
 
 | | Structure of RS223-Beta-xylosidase | | Descriptor: | Beta-xylosidase, CALCIUM ION, SULFATE ION | | Authors: | Jordan, D, Braker, J, Wagschal, K, Lee, C, Dubrovska, I, Anderson, S, Wawrzak, Z. | | Deposit date: | 2013-09-06 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of RS223-Beta-xylosidase
To be Published
|
|
3OSU
 
 | | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Edwards, A, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-09 | | Release date: | 2010-09-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the 3-oxoacyl-acyl carrier protein reductase, FabG, from Staphylococcus aureus
To be Published
|
|
3OYT
 
 | | 1.84 Angstrom resolution crystal structure of 3-oxoacyl-(acyl carrier protein) synthase I (fabB) from Yersinia pestis CO92 | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase I, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-23 | | Release date: | 2011-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | 1.84 Angstrom resolution crystal structure of 3-oxoacyl-(acyl carrier protein) synthase I (fabB) from Yersinia pestis CO92
To be Published
|
|
3OJC
 
 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
3OUT
 
 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-09-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
3P0R
 
 | | Crystal structure of azoreductase from Bacillus anthracis str. Sterne | | Descriptor: | Azoreductase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of azoreductase from Bacillus anthracis str. Sterne
To be Published
|
|
3P52
 
 | | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NITRATE ION | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion
To be Published
|
|
4N3O
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni. | | Descriptor: | CALCIUM ION, Putative D-glycero-D-manno-heptose 7-phosphate kinase | | Authors: | Minasov, G, Wawrzak, Z, Gordon, E, Onopriyenko, O, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-07 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni.
TO BE PUBLISHED
|
|
3H02
 
 | | 2.15 Angstrom Resolution Crystal Structure of Naphthoate Synthase from Salmonella typhimurium. | | Descriptor: | BICARBONATE ION, Naphthoate synthase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Naphthoate Synthase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3H0P
 
 | | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium. | | Descriptor: | S-malonyltransferase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3GRI
 
 | | The Crystal Structure of a Dihydroorotase from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Dihydroorotase from Staphylococcus aureus
To be Published
|
|
3GSD
 
 | | 2.05 Angstrom structure of a divalent-cation tolerance protein (CutA) from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Structure of a Divalent-cation Tolerance Protein (CutA) from Yersinia pestis
TO BE PUBLISHED
|
|
3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3IAC
 
 | | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium. | | Descriptor: | CHLORIDE ION, Glucuronate isomerase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium.
To be Published
|
|
3IFS
 
 | | 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase, LITHIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Gordon, E, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis.
To be Published
|
|
3IG4
 
 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
3IMI
 
 | | 2.01 Angstrom resolution crystal structure of a HIT family protein from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | HIT family protein, SULFATE ION, ZINC ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Gordon, E, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2.01 Angstrom resolution crystal structure of a HIT family protein from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3IGX
 
 | | 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis. | | Descriptor: | PHOSPHATE ION, Transaldolase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Gordon, E, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis.
TO BE PUBLISHED
|
|
3K1O
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Trypanosoma cruzi in complex with a potential antichagasic drug, posaconazole | | Descriptor: | 2,5-anhydro-1,3,4-trideoxy-2-(2,4-difluorophenyl)-6-O-{4-[4-(4-{1-[(1S,2S)-1-ethyl-2-hydroxypropyl]-5-oxo-1,5-dihydro-4H-1,2,4-triazol-4-yl}phenyl)piperazin-1-yl]phenyl}-1-(1H-1,2,4-triazol-1-yl)-D-erythro-hexitol, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14 alpha-demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Anderson, S, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Insights into Inhibition of Sterol 14{alpha}-Demethylase in the Human Pathogen Trypanosoma cruzi.
J.Biol.Chem., 285, 2010
|
|
3I3W
 
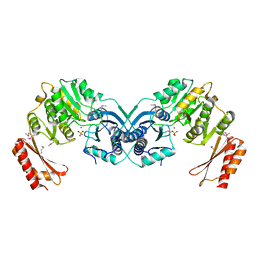 | | Structure of a phosphoglucosamine mutase from Francisella tularensis | | Descriptor: | Phosphoglucosamine mutase, ZINC ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a phosphoglucosamine mutase from Francisella tularensis
To be Published
|
|
3K28
 
 | | Crystal Structure of a glutamate-1-semialdehyde aminotransferase from Bacillus anthracis with bound Pyridoxal 5'Phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glutamate-1-semialdehyde 2,1-aminomutase 2, ... | | Authors: | Sharma, S.S, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-29 | | Release date: | 2010-01-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a glutamate-1-semialdehyde aminotransferase from Bacillus anthracis with bound Pyridoxal 5'Phosphate
To be Published
|
|
3GW9
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei bound to an inhibitor N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxaziazol-2-yl)benzamide | | Descriptor: | N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14ALPHA-DEMETHYLASE | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Harp, J, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections.
J.Biol.Chem., 285, 2010
|
|
