6KB6
 
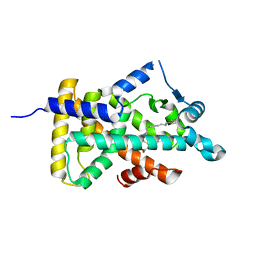 | | X-ray structure of human PPARalpha ligand binding domain-tetradecylthioacetic acid (TTA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-tetradecylsulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.431 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L1A
 
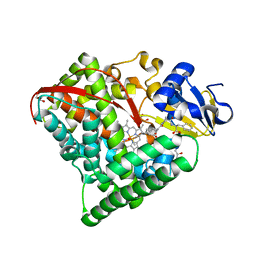 | | Crystal Structure of P450BM3 with N-enanthoyl-L-prolyl-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-heptanoylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Shoji, O, Yonemura, K. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
6KYP
 
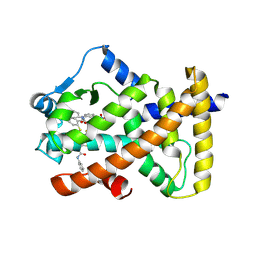 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-clofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-09-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L37
 
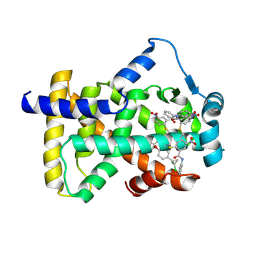 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
2Z01
 
 | | Crystal structure of phosphoribosylaminoimidazole synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 2015
|
|
6L1B
 
 | | Crystal Structure of P450BM3 with N-(3-cyclopentylpropanoyl)-L-pipecolyl-L-phenylalanine | | Descriptor: | (2S)-2-[[(2S)-1-(3-cyclopentylpropanoyl)piperidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Shoji, O, Yonemura, K. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
6L36
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L38
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-gemfibrozil co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.761 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX4
 
 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX5
 
 | | X-ray structure of human PPARalpha ligand binding domain-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX6
 
 | | X-ray structure of human PPARalpha ligand binding domain-palmitic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXB
 
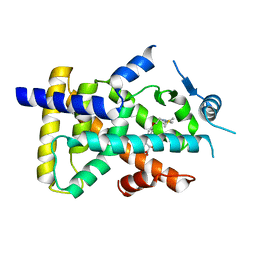 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by soaking | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXA
 
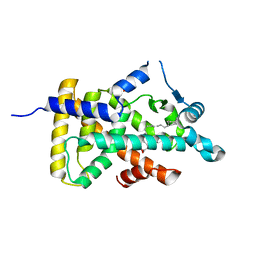 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX8
 
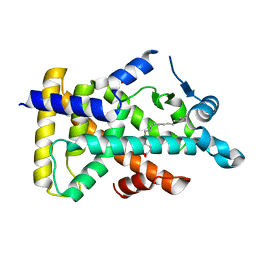 | | X-ray structure of human PPARalpha ligand binding domain-oleic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, OLEIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX7
 
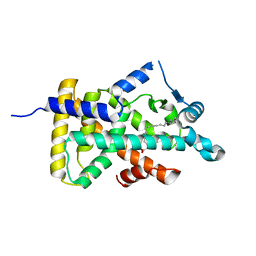 | | X-ray structure of human PPARalpha ligand binding domain-stearic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, STEARIC ACID | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXC
 
 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by delipidation and cross-seeding | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6JAF
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with PPi (pyrophosphatase reaction) | | Descriptor: | GLYCEROL, Glycerol kinase, PYROPHOSPHATE 2- | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, Bringaud, F, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glycerol Kinase of African Human Trypanosomes Possesses a Pyrophosphatase Activity.
To Be Published
|
|
6J9X
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase phosphorylated at Thr12(pyrophosphatase reaction) | | Descriptor: | GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, Bringaud, F, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Glycerol Kinase of African Human Trypanosomes Possesses a Pyrophosphatase Activity.
To Be Published
|
|
6JAE
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with Pi (pyrophosphatase reaction) | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHATE ION | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, Bringaud, F, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycerol Kinase of African Human Trypanosomes Possesses a Pyrophosphatase Activity.
To Be Published
|
|
