5HNQ
 
 | |
5HN2
 
 | |
5HNJ
 
 | |
5XP6
 
 | | native structure of NDM-1 crystallized at pH5.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G, Lai, J, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
5XP9
 
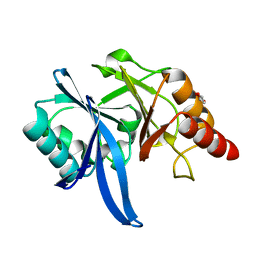 | | Crystal structure of Bismuth bound NDM-1 | | Descriptor: | Bismuth(III) ION, GLYCEROL, Metallo-beta-lactamase type 2 | | Authors: | Zhang, H, Ma, G, Lai, T.P, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
2L8E
 
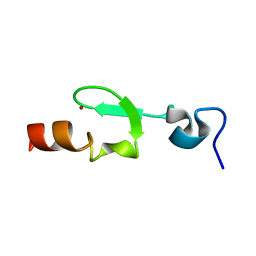 | |
8VB1
 
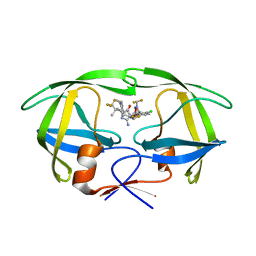 | | Crystal structure of HIV-1 protease with GS-9770 | | Descriptor: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | Authors: | Lansdon, E.B. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
8X84
 
 | |
8X83
 
 | |
8X82
 
 | |
4XGY
 
 | | GFP based antibody (fluorobody) | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Green fluorescent protein, mAb LCDR3 | | Authors: | Shi, N, Chen, Y.G, Wang, S.H. | | Deposit date: | 2015-01-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | The structure of a GFP-based antibody (fluorobody) to TLH, a toxin from Vibrio parahaemolyticus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6C6P
 
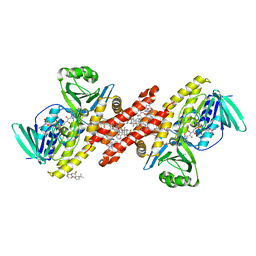 | | Human squalene epoxidase (SQLE, squalene monooxygenase) structure with FAD and NB-598 | | Descriptor: | (2E)-N-({3-[([3,3'-bithiophen]-5-yl)methoxy]phenyl}methyl)-N-ethyl-6,6-dimethylhept-2-en-4-yn-1-amine, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Padyana, A.K, Jin, L. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase.
Nat Commun, 10, 2019
|
|
6C6N
 
 | |
6C6R
 
 | |
6LAD
 
 | |
6LAF
 
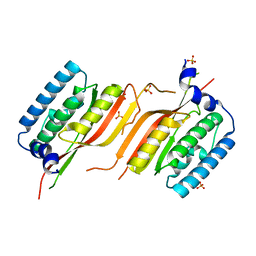 | | Crystal structure of the core domain of Amuc_1100 from Akkermansia muciniphila | | Descriptor: | Amuc_1100, SULFATE ION | | Authors: | Wang, J, Xiang, R, Zhang, M, Wang, M. | | Deposit date: | 2019-11-12 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | The variable oligomeric state of Amuc_1100 from Akkermansia muciniphila.
J.Struct.Biol., 212, 2020
|
|
7KZP
 
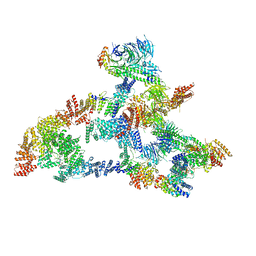 | | Structure of the human Fanconi anaemia Core complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KZT
 
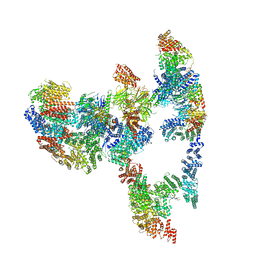 | |
7KZV
 
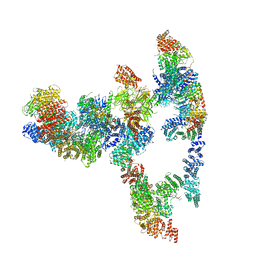 | |
7KZQ
 
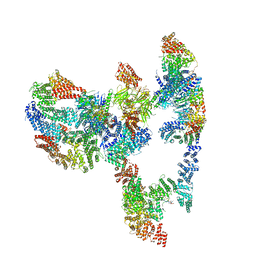 | | Structure of the human Fanconi anaemia Core-ID complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KZR
 
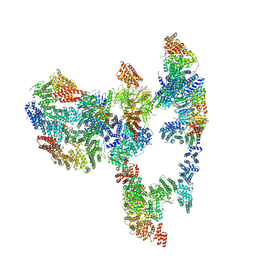 | | Structure of the human Fanconi Anaemia Core-UBE2T-ID complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KZS
 
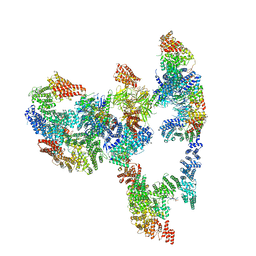 | |
5UEW
 
 | | BRD2 Bromodomain2 with A-1360579 | | Descriptor: | Bromodomain-containing protein 2, N-[3-(4-methoxy-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEY
 
 | | BRD4_BD2_A-1412838 | | Descriptor: | 5-[2-(2,4-difluorophenoxy)-5-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}phenyl]-4-ethoxy-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEX
 
 | | BRD4_BD2_A-1497627 | | Descriptor: | 17-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}-11,13-difluoro-2-methyl-6,7,8,9-tetrahydrodibenzo[4,5:7,8][1,6]dioxacyclododecino[3,2-c]pyridin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
