5FB5
 
 | |
5FEI
 
 | | Crystal structure of the bacteriophage phi29 tail knob protein gp9 truncation variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Distal tube protein | | Authors: | Xu, J.W, Gui, M, Wang, D.H, Xiang, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The bacteriophage 29 tail possesses a pore-forming loop for cell membrane penetration.
Nature, 534, 2016
|
|
5FB4
 
 | | Crystal structure of the bacteriophage phi29 tail knob protein gp9 truncation variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Distal tube protein | | Authors: | Xu, J.W, Gui, M, Wang, D.H, Xiang, Y. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | The bacteriophage 29 tail possesses a pore-forming loop for cell membrane penetration.
Nature, 534, 2016
|
|
4IN4
 
 | | Crystal structure of cpd 15 bound to Keap1 Kelch domain | | Descriptor: | 2-({5-[(2,4-dimethylphenyl)sulfonyl]-6-oxo-1,6-dihydropyrimidin-2-yl}sulfanyl)-N-[2-(trifluoromethyl)phenyl]acetamide, Kelch-like ECH-associated protein 1, PHOSPHATE ION | | Authors: | Silvian, L, Marcotte, D. | | Deposit date: | 2013-01-03 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism.
Bioorg.Med.Chem., 21, 2013
|
|
6LLG
 
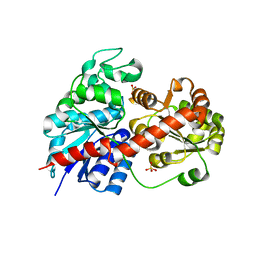 | | Crystal Structure of Fagopyrum esculentum M UGT708C1 | | Descriptor: | BENZAMIDINE, SULFATE ION, UDP-glycosyltransferase 708C1 | | Authors: | Wang, X, Liu, M. | | Deposit date: | 2019-12-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of theC-Glycosyltransferase UGT708C1 from Buckwheat Provide Insights into the Mechanism ofC-Glycosylation.
Plant Cell, 32, 2020
|
|
6LLZ
 
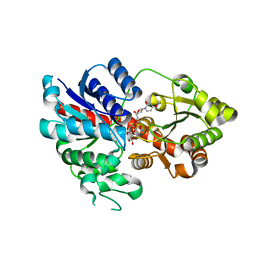 | |
4IQK
 
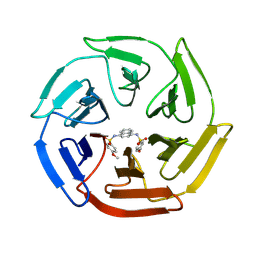 | | Crystal structure of cpd 16 bound to Keap1 Kelch domain | | Descriptor: | Kelch-like ECH-associated protein 1, N,N'-naphthalene-1,4-diylbis(4-methoxybenzenesulfonamide) | | Authors: | Silvian, L, Marcotte, D. | | Deposit date: | 2013-01-11 | | Release date: | 2013-05-15 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism.
Bioorg.Med.Chem., 21, 2013
|
|
6LLW
 
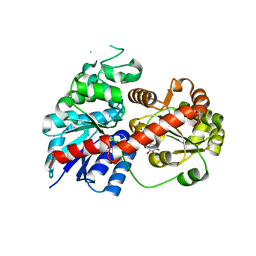 | |
4R6U
 
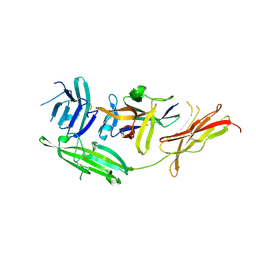 | | IL-18 receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-18, Interleukin-18 receptor 1 | | Authors: | Wei, H, Wang, X. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the specific recognition of IL-18 by its alpha receptor.
Febs Lett., 588, 2014
|
|
4R99
 
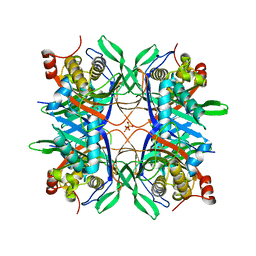 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | SULFATE ION, Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
2OYS
 
 | | Crystal Structure of SP1951 protein from Streptococcus pneumoniae in complex with FMN, Northeast Structural Genomics Target SpR27 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Hypothetical protein SP1951 | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Janjua, H, Satterwhite, R, Liu, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
6MG1
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta, ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MG3
 
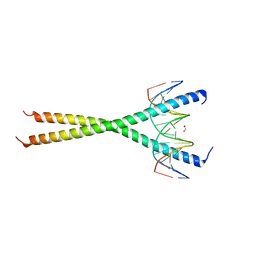 | | V285A Mutant of the C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MG2
 
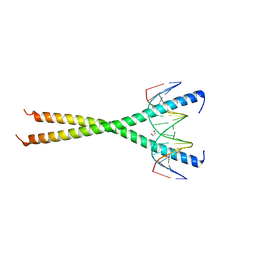 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2221 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6LQZ
 
 | | Solution structure of Taf14ET-Sth1EBMC | | Descriptor: | Nuclear protein STH1/NPS1, Transcription initiation factor TFIID subunit 14 | | Authors: | Wu, B, Chen, G, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation.
Nat Commun, 11, 2020
|
|
5U5Q
 
 | | 12 Subunit RNA Polymerase II at Room Temperature collected using SFX | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Bushnell, D.A, Oberthur, D, Mariani, V, Yefanov, O, Tolstikova, A, Barty, A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Double-flow focused liquid injector for efficient serial femtosecond crystallography.
Sci Rep, 7, 2017
|
|
8F1D
 
 | | Voltage-gated potassium channel Kv3.1 apo | | Descriptor: | CHOLESTEROL, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1C
 
 | | Voltage-gated potassium channel Kv3.1 with novel positive modulator (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one (compound 4) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one, POTASSIUM ION, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JGA
 
 | | Cryo-EM structure of Mi3 fused with FKBP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8JGC
 
 | | Cryo-EM structure of Mi3 fused with LOV2 | | Descriptor: | LOV domain-containing protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
1ZBP
 
 | | X-Ray Crystal Structure of Protein VPA1032 from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR44 | | Descriptor: | hypothetical protein VPA1032 | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Ciao, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
6OUS
 
 | | Structure of fusion glycoprotein from human respiratory syncytial virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, ... | | Authors: | Su, H.P. | | Deposit date: | 2019-05-05 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A potent broadly neutralizing human RSV antibody targets conserved site IV of the fusion glycoprotein.
Nat Commun, 10, 2019
|
|
6CF1
 
 | | Proteus vulgaris HigA antitoxin structure | | Descriptor: | Antitoxin HigA, POTASSIUM ION | | Authors: | Schureck, M.A, Hoffer, E.D, Ei Cho, S, Dunham, C.M. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
6CHV
 
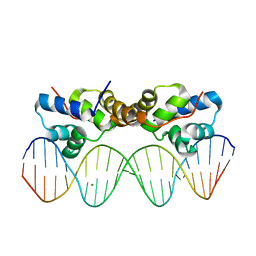 | | Proteus vulgaris HigA antitoxin bound to DNA | | Descriptor: | Antitoxin HigA, MAGNESIUM ION, pHigCryst3, ... | | Authors: | Schureck, M.A, Hoffer, E.D, Onuoha, N, Dunham, C.M. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
