4M7C
 
 | | Crystal structure of the TRF2-binding motif of SLX4 in complex with the TRFH domain of TRF2 | | Descriptor: | Peptide from Structure-specific endonuclease subunit SLX4, Telomeric repeat-binding factor 2 | | Authors: | Wan, B, Chen, Y, Wu, J, Liu, Y, Lei, M. | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SLX4 Assembles a Telomere Maintenance Toolkit by Bridging Multiple Endonucleases with Telomeres
Cell Rep, 4, 2013
|
|
4ZOU
 
 | |
5DOI
 
 | | Crystal structure of Tetrahymena p45N and p19 | | Descriptor: | Telomerase associated protein p45, Telomerase-associated protein 19 | | Authors: | Wan, B, Tang, T, Wu, J, Lei, M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DOF
 
 | | Crystal structure of Tetrahymena p19 | | Descriptor: | Telomerase-associated protein 19 | | Authors: | Wan, B, Tang, T, Wu, J, Lei, M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DOK
 
 | | Crystal structure of Tetrahymena p45C | | Descriptor: | MAGNESIUM ION, Telomerase associated protein p45 | | Authors: | Wan, B, Tang, T, Wu, J, Lei, M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
6J0W
 
 | | Crystal Structure of Yeast Rtt107 and Nse6 | | Descriptor: | Peptide from DNA repair protein KRE29, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0V
 
 | | Crystal Structure of Yeast Rtt107 | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0Y
 
 | | Crystal Structure of Yeast Rtt107 and Slx4 | | Descriptor: | Peptide from Structure-specific endonuclease subunit SLX4, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0X
 
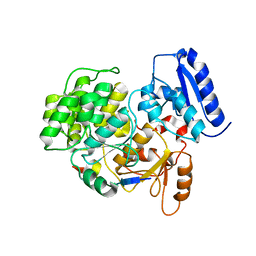 | | Crystal Structure of Yeast Rtt107 and Mms22 | | Descriptor: | Peptide from E3 ubiquitin-protein ligase substrate receptor MMS22, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
7BU5
 
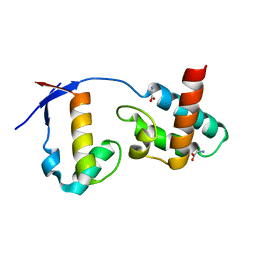 | | Crystal Structure of Human SLX4 and MUS81 | | Descriptor: | GLYCINE, MUS81 endonuclease homolog (Yeast), isoform CRA_b, ... | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SLX4 control MUS81 endonuclease function in telomere through an intermolecular SAP domain
To be published
|
|
8GNH
 
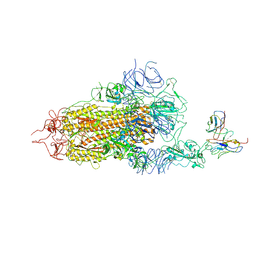 | | Complex structure of BD-218 and Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BD-218, ... | | Authors: | Wang, B, Xu, H, Su, X.D. | | Deposit date: | 2022-08-23 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Human antibody BD-218 has broad neutralizing activity against concerning variants of SARS-CoV-2.
Int.J.Biol.Macromol., 227, 2023
|
|
6KKS
 
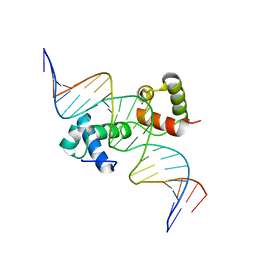 | | Structural insights into target DNA recognition by R2R3-type MYB transcription factor | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*CP*TP*CP*CP*AP*AP*CP*CP*GP*CP*AP*TP*TP*TP*TP*C)-3'), DNA (5'-D(*CP*GP*AP*AP*AP*AP*TP*GP*CP*GP*GP*TP*TP*GP*GP*AP*GP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Wang, B, Luo, Q. | | Deposit date: | 2019-07-27 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into target DNA recognition by R2R3-MYB transcription factors.
Nucleic Acids Res., 48, 2020
|
|
1NJ3
 
 | | Structure and Ubiquitin Interactions of the Conserved NZF Domain of Npl4 | | Descriptor: | NPL4, ZINC ION | | Authors: | Wang, B, Alam, S.L, Meyer, H.H, Payne, M, Stemmler, T.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4.
J.Biol.Chem., 278, 2003
|
|
6P3W
 
 | |
5J62
 
 | | FMN-dependent Nitroreductase (CDR20291_0684) from Clostridium difficile R20291 | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Wang, B, Powell, S.M, Hessami, N, Najar, F.Z, Thomas, L.M, West, A.H, Karr, E.A, Richter-Addo, G.B. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of two nitroreductases from hypervirulent Clostridium difficile and functionally related interactions with the antibiotic metronidazole.
Nitric Oxide, 60, 2016
|
|
1FFP
 
 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH PEPTIDE GP33 (C9M/K1S) | | Descriptor: | BETA-2 MICROGLOBULIN BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition
J.IMMUNOL., 169, 2002
|
|
1FFN
 
 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH PEPTIDE GP33(C9M) | | Descriptor: | BETA-2 MICROGLOBULIN, BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition
J.IMMUNOL., 169, 2002
|
|
1FFO
 
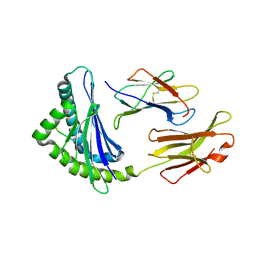 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH SYNTHETIC PEPTIDE GP33 (C9M/K1A) | | Descriptor: | BETA-2 MICROGLOBULIN BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition.
J.Immunol., 169, 2002
|
|
5KD1
 
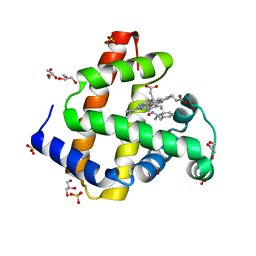 | | Sperm whale myoglobin H64A with nitrosoamphetamine | | Descriptor: | GLYCEROL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, B, Guan, Y, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2016-06-07 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nitrosoamphetamine binding to myoglobin and hemoglobin: Crystal structure of the H64A myoglobin-nitrosoamphetamine adduct.
Nitric Oxide, 67, 2017
|
|
5B1Q
 
 | | Human herpesvirus 6B tegument protein U14 | | Descriptor: | GLYCEROL, U14 protein | | Authors: | Wang, B, Nishimura, M, Tang, H, Kawabata, A, Mahmoud, N.F, Khanlari, Z, Hamada, D, Tsuruta, H, Mori, Y. | | Deposit date: | 2015-12-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Human Herpesvirus 6B Tegument Protein U14.
Plos Pathog., 12, 2016
|
|
1QYP
 
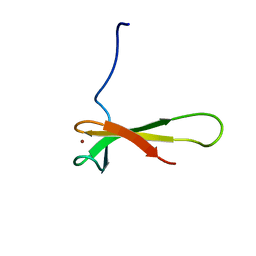 | | THERMOCOCCUS CELER RPB9, NMR, 25 STRUCTURES | | Descriptor: | RNA POLYMERASE II, ZINC ION | | Authors: | Wang, B, Jones, D.N.M, Kaine, B.P, Weiss, M.A. | | Deposit date: | 1997-08-19 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an archaeal zinc ribbon defines a general architectural motif in eukaryotic RNA polymerases.
Structure, 6, 1998
|
|
6UX2
 
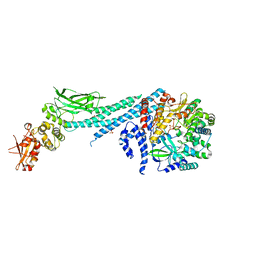 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6CZF
 
 | | The structure of E. coli PurF in complex with ppGpp-Mg | | Descriptor: | Amidophosphoribosyltransferase, GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Affinity-based capture and identification of protein effectors of the growth regulator ppGpp.
Nat. Chem. Biol., 15, 2019
|
|
6VWP
 
 | | Crystal structure of E. coli guanosine kinase in complex with ppGpp | | Descriptor: | GUANOSINE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-guanosine kinase, ... | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | ppGpp Coordinates Nucleotide and Amino-Acid Synthesis in E. coli During Starvation.
Mol.Cell, 80, 2020
|
|
6VWO
 
 | | Crystal structure of E. coli guanosine kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE, Inosine-guanosine kinase, ... | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | ppGpp Coordinates Nucleotide and Amino-Acid Synthesis in E. coli During Starvation.
Mol.Cell, 80, 2020
|
|
