438D
 
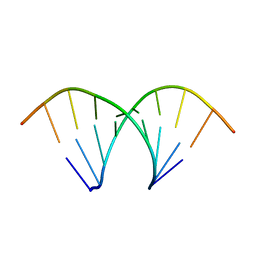 | |
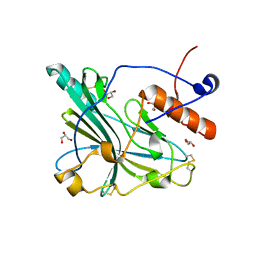
6SQJ
 
 | | Crystal structure of glycoprotein D of Equine Herpesvirus Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein D, ... | | Authors: | Kremling, V, Loll, B, Azab, W, Osterrieder, N, Dahmani, I, Chiantia, P, Wahl, M. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Crystal structures of glycoprotein D of equine alphaherpesviruses reveal potential binding sites to the entry receptor MHC-I.
Front Microbiol, 14, 2023
|
|
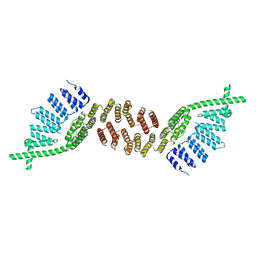
6TM8
 
 | | Crystal structure of glycoprotein D of Equine Herpesvirus Type 4 | | Descriptor: | Envelope glycoprotein D, GLYCEROL | | Authors: | Kremling, V, Loll, B, Osterrieder, N, Wahl, M, Dahmani, I, Chiantia, P, Azab, W. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycoprotein D of equine alphaherpesviruses reveal potential binding sites to the entry receptor MHC-I.
Front Microbiol, 14, 2023
|
|
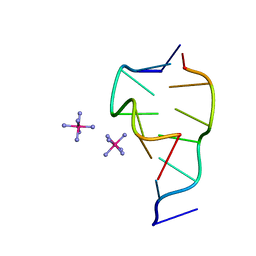
6G70
 
 | | Structure of murine Prpf39 | | Descriptor: | Pre-mRNA-processing factor 39 | | Authors: | De Bortoli, F.D, Loll, B, Wahl, M, Heyd, F. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Increased versatility despite reduced molecular complexity: evolution, structure and function of metazoan splicing factor PRPF39.
Nucleic Acids Res., 47, 2019
|
|
331D
 
 | | CRYSTAL STRUCTURE OF D(GCGCGCG) WITH 5'-OVERHANG G'S | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*GP*CP*GP*CP*G)-3') | | Authors: | Pan, B, Ban, C, Wahl, M, Sundaralingam, M. | | Deposit date: | 1997-05-13 | | Release date: | 1997-09-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of d(GCGCGCG) with 5'-overhang G residues.
Biophys.J., 73, 1997
|
|
7ADC
 
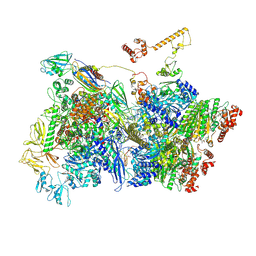 | | Transcription termination intermediate complex 3 delta NusG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADB
 
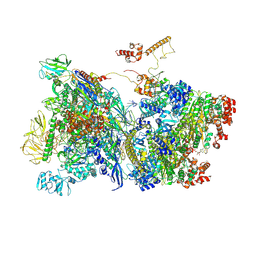 | | Transcription termination intermediate complex 1 delta NusG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADD
 
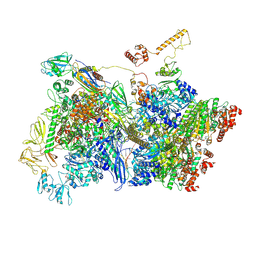 | | Transcription termination intermediate complex IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADE
 
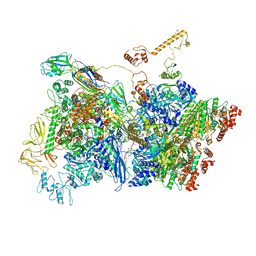 | | Transcription termination complex IVa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7O6Q
 
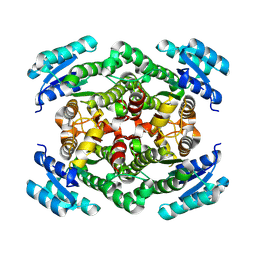 | |
7O6P
 
 | |
5MS0
 
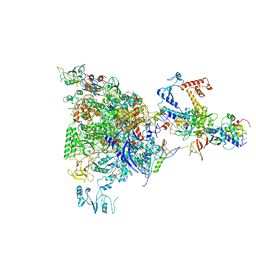 | |
5JDT
 
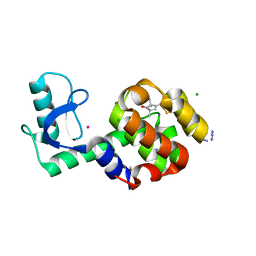 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at 100K | | Descriptor: | AZIDE ION, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-04-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
7Z3S
 
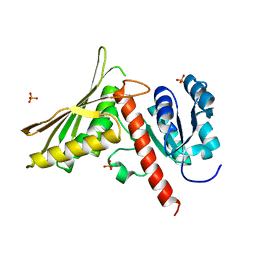 | |
2KVQ
 
 | |
1GC2
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1GC0
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1NPR
 
 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN C222(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzhan, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
1NPP
 
 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN P2(1) | | Descriptor: | ISOPROPYL ALCOHOL, Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzahn, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
7PH1
 
 | | Trypsin in complex with BPTI mutant (2S)-2-amino-4-monofluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Loll, B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Water Network in the Binding Pocket of Fluorinated BPTI-Trypsin Complexes─Insights from Simulation and Experiment.
J.Phys.Chem.B, 126, 2022
|
|
7QIQ
 
 | | CRYSTAL STRUCTURE OF THE P1 aminobutanoic acid (ABU) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIT
 
 | | CRYSTAL STRUCTURE OF THE P1 trifluoroethylglycine (TfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIS
 
 | | CRYSTAL STRUCTURE OF THE P1 difluoroethylglycine (DfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIR
 
 | | CRYSTAL STRUCTURE OF THE P1 monofluorethylglycine(MfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
4PKD
 
 | | U1-70k in complex with U1 snRNA stem-loops 1 and U1-A RRM in complex with stem-loop 2 | | Descriptor: | IMIDAZOLE, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A,U1 small nuclear ribonucleoprotein 70 kDa, ... | | Authors: | Oubridge, C, Kondo, Y, van Roon, A.M, Nagai, K. | | Deposit date: | 2014-05-14 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition.
Elife, 4, 2015
|
|
