6IIG
 
 | |
6KCJ
 
 | |
3EZJ
 
 | | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody | | Descriptor: | CHLORIDE ION, General secretion pathway protein GspD, NANOBODY NBGSPD_7, ... | | Authors: | Korotkov, K.V, Pardon, E, Steyaert, J, Hol, W.G. | | Deposit date: | 2008-10-22 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody.
Structure, 17, 2009
|
|
6LZH
 
 | | Crystal structure of Alpha/beta hydrolase GrgF from Penicillium sp. sh18 | | Descriptor: | GrgF, SODIUM ION | | Authors: | Wang, H, Yu, J, Wang, W.G, Matsuda, Y, Yao, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Biosynthesis of an Unusual Chain-Fused Polyketide, Gregatin A.
J.Am.Chem.Soc., 142, 2020
|
|
6AZH
 
 | |
1A35
 
 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1B1B
 
 | | IRON DEPENDENT REGULATOR | | Descriptor: | PROTEIN (IRON DEPENDENT REGULATOR), SULFATE ION, ZINC ION | | Authors: | Pohl, E, Holmes, R.K, Hol, W.G. | | Deposit date: | 1998-11-19 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied.
J.Mol.Biol., 285, 1999
|
|
1B5S
 
 | | DIHYDROLIPOYL TRANSACETYLASE (E.C.2.3.1.12) CATALYTIC DOMAIN (RESIDUES 184-425) FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Izard, T, Aevarsson, A, Allen, M.D, Westphal, A.H, Perham, R.N, De Kok, A, Hol, W.G. | | Deposit date: | 1999-01-10 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C0W
 
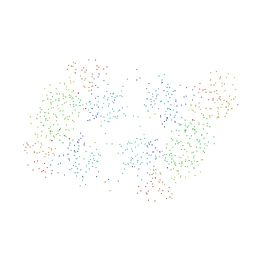 | | CRYSTAL STRUCTURE OF THE COBALT-ACTIVATED DIPHTHERIA TOXIN REPRESSOR-DNA COMPLEX REVEALS A METAL BINDING SH-LIKE DOMAIN | | Descriptor: | COBALT (II) ION, DIPHTHERIA TOXIN REPRESSOR, DNA (5'-D(P*AP*TP*TP*AP*GP*GP*TP*TP*AP*GP*CP*CP*TP*AP*CP*CP*CP*TP*AP*AP*T)-3'), ... | | Authors: | Pohl, E, Holmes, R.K, Hol, W.G. | | Deposit date: | 1999-07-22 | | Release date: | 2000-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain.
J.Mol.Biol., 292, 1999
|
|
1RQC
 
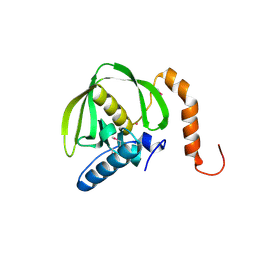 | | Crystals of peptide deformylase from Plasmodium falciparum with ten subunits per asymmetric unit reveal critical characteristics of the active site for drug design | | Descriptor: | COBALT (II) ION, formylmethionine deformylase | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G. | | Deposit date: | 2003-12-04 | | Release date: | 2004-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase
Protein Sci., 13, 2004
|
|
1TV6
 
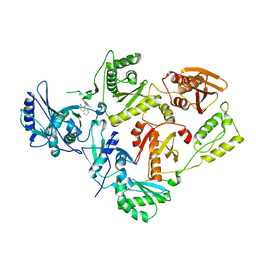 | | HIV-1 Reverse Transcriptase Complexed with CP-94,707 | | Descriptor: | 3-[4-(2-METHYL-IMIDAZO[4,5-C]PYRIDIN-1-YL)BENZYL]-3H-BENZOTHIAZOL-2-ONE, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit | | Authors: | Pata, J.D, Stirtan, W.G, Goldstein, S.W, Steitz, T.A. | | Deposit date: | 2004-06-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant RTs resistant to other non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2OEU
 
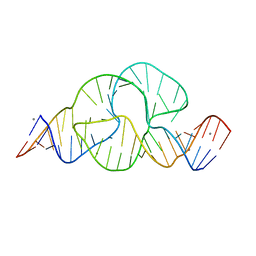 | | Full-length hammerhead ribozyme with Mn(II) bound | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*(OMC)P*CP*UP*GP*GP*(5BU)P*AP*UP*CP*CP*AP*AP*UP*CP*(DC))-3', Hammerhead Ribozyme, MANGANESE (II) ION | | Authors: | Martick, M, Scott, W.G. | | Deposit date: | 2007-01-01 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solvent structure and hammerhead ribozyme catalysis.
Chem.Biol., 15, 2008
|
|
2OIU
 
 | | L1 Ribozyme Ligase circular adduct | | Descriptor: | L1 Ribozyme RNA Ligase, MAGNESIUM ION | | Authors: | Robertson, M.P, Scott, W.G. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of ribozyme-catalyzed RNA assembly.
Science, 315, 2007
|
|
2PMW
 
 | | The Crystal Structure of Proprotein convertase subtilisin kexin type 9 (PCSK9) | | Descriptor: | Proprotein convertase subtilisin/kexin type 9, SULFATE ION | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of PCSK9: A Regulator of Plasma LDL-Cholesterol.
Structure, 15, 2007
|
|
2QUS
 
 | |
2QUW
 
 | |
2VMK
 
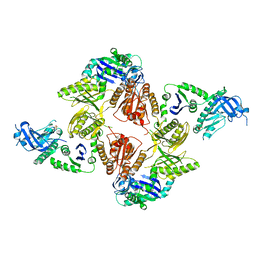 | | Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain | | Descriptor: | RIBONUCLEASE E, SULFATE ION, ZINC ION | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
2AEP
 
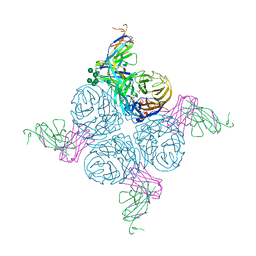 | | An epidemiologically significant epitope of a 1998 influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FAB heavy chain, ... | | Authors: | Venkatramani, L, Bochkareva, E, Lee, J.T, Gulati, U, Laver, W.G, Bochkarev, A, Air, G.M. | | Deposit date: | 2005-07-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Epidemiologically Significant Epitope of a 1998 Human Influenza Virus Neuraminidase Forms a Highly Hydrated Interface in the NA-Antibody Complex
J.Mol.Biol., 356, 2006
|
|
299D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: THE HAMMERHEAD RIBOZYME | | Descriptor: | RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
2AF6
 
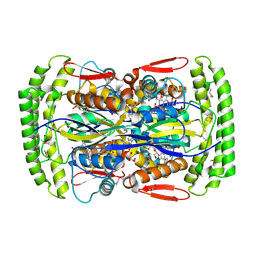 | | Crystal structure of Mycobacterium tuberculosis Flavin dependent thymidylate synthase (Mtb ThyX) in the presence of co-factor FAD and substrate analog 5-Bromo-2'-Deoxyuridine-5'-Monophosphate (BrdUMP) | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Sampathkumar, P, Turley, S, Ulmer, J.E, Rhie, H.G, Sibley, C.H, Hol, W.G. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Mycobacterium tuberculosis Flavin Dependent Thymidylate Synthase (MtbThyX) at 2.0A Resolution.
J.Mol.Biol., 352, 2005
|
|
2B51
 
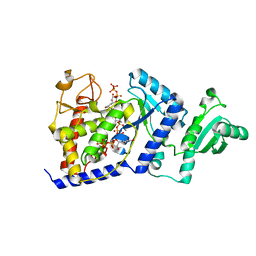 | | Structural Basis for UTP Specificity of RNA Editing TUTases from Trypanosoma Brucei | | Descriptor: | MANGANESE (II) ION, RNA editing complex protein MP57, URIDINE 5'-TRIPHOSPHATE | | Authors: | Deng, J, Ernst, N.L, Turley, S, Stuart, K.D, Hol, W.G. | | Deposit date: | 2005-09-27 | | Release date: | 2005-11-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
Embo J., 24, 2005
|
|
2B4V
 
 | | Structural Basis for UTP Specificity of RNA Editing TUTases From Trypanosoma Brucei | | Descriptor: | POTASSIUM ION, RNA editing complex protein MP57 | | Authors: | Deng, J, Ernst, N.L, Turley, S, Stuart, K.D, Hol, W.G. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
Embo J., 24, 2005
|
|
2B56
 
 | | Structural Basis for UTP Specificity of RNA Editing TUTases From Trypanosoma Brucei | | Descriptor: | MAGNESIUM ION, RNA editing complex protein MP57, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Deng, J, Ernst, N.L, Turley, S, Stuart, K.D, Hol, W.G. | | Deposit date: | 2005-09-27 | | Release date: | 2005-11-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
Embo J., 24, 2005
|
|
2C4A
 
 | | Structure of Neuraminidase Subtype N9 Complexed with 30 MM Sialic Acid (NANA, NEU5AC), Crystal Soaked for 3 Hours at 291 K. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2005-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Influenza Type a Virus Neuraminidase of the N6 Subtype at 1.85 A Resolution
To be Published
|
|
2C4L
 
 | | Structure of Neuraminidase Subtype N9 Complexed with 30 MM Sialic Acid (NANA, NEU5AC), Crystal Soaked for 24 Hours at 291 K and Finally Backsoaked for 30 Min in a Cryoprotectant Solution which did not contain NEU5AC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2005-10-20 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Influenza Type a Virus Neuraminidase of the N6 Subtype at 1.85 A Resolution
To be Published
|
|
